Abstract
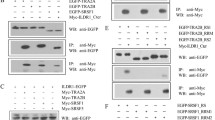
Fas is a transmembrane cell surface protein recognized by Fas ligand (FasL). When FasL binds to Fas, the target cells undergo apoptosis. A soluble Fas molecule that lacks the transmembrane domain is produced from skipping of exon 6 encoding this region in alternative splicing procedure. The soluble Fas molecule has the opposite function of intact Fas molecule, protecting cells from apoptosis. Here we show that knockdown of hnRNP A1 promotes exon 6 skipping of Fas pre-mRNA, whereas overexpression of hnRNP A1 reduces exon 6 skipping. Based on the bioinformatics approach, we have hypothesized that hnRNP A1 functions through interrupting 5′ splice site selection of exon 5 by interacting with its potential binding site close to 5′ splice site of exon 5. Consistent with our hypothesis, we demonstrate that mutations of the hnRNP A1 binding site on exon 5 disrupted the effects of hnRNP A1 on exon 6 inclusion. RNA pull-down assay and then western blot analysis with hnRNP A1 antibody prove that hnRNP A1 contacts the potential binding site RNA sequence on exon 5 but not the mutant sequence. In addition, we show that the mutation of 5′ splice site on exon 5 to a less conserved sequence destructed the effects of hnRNP A1 on exon 6 inclusion. Therefore we conclude that hnRNP A1 interacts with exon 5 to promote distal exon 6 inclusion of Fas pre-mRNA. Our study reveals a novel alternative splicing mechanism of Fas pre-mRNA.






Similar content being viewed by others
References
Itoh N, Yonehara S, Ishii A, Yonehara M, Mizushima S, Sameshima M et al (1991) The polypeptide encoded by the cDNA for human cell surface antigen Fas can mediate apoptosis. Cell 66(2):233–243
Trauth BC, Klas C, Peters AM, Matzku S, Moller P, Falk W et al (1989) Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science 245(4915):301–305
Oehm A, Behrmann I, Falk W, Pawlita M, Maier G, Klas C et al (1992) Purification and molecular cloning of the APO-1 cell surface antigen, a member of the tumor necrosis factor/nerve growth factor receptor superfamily. Sequence identity with the Fas antigen. J Biol Chem 267(15):10709–10715
Lee SH, Kim SY, Lee JY, Shin MS, Dong SM, Na EY et al (1998) Detection of soluble Fas mRNA using in situ reverse transcription-polymerase chain reaction. Lab Invest 78(4):453–459
Wahl MC, Will CL, Luhrmann R (2009) The spliceosome: Design principles of a dynamic RNP machine. Cell 136(4):701–718. doi:10.1016/j.cell.2009.02.009
Black DL (2000) Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell 103(3):367–370
Graveley BR (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet 17(2):100–107
Modrek B, Resch A, Grasso C, Lee C (2001) Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res 29(13):2850–2859
Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72:291–336. doi:10.1146/annurev.biochem.72.121801.161720
Matlin AJ, Clark F, Smith CW (2005) Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol 6(5):386–398. doi:10.1038/nrm1645
Jurica MS, Moore MJ (2003) Pre-mRNA splicing: awash in a sea of proteins. Mol Cell 12(1):5–14
Reed R (1996) Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Genet Dev 6(2):215–220
Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG (1993) hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem 62:289–321. doi:10.1146/annurev.bi.62.070193.001445
Hertel KJ (2008) Combinatorial control of exon recognition. J Biol Chem 283(3):1211–1215. doi:10.1074/jbc.R700035200
Senapathy P, Shapiro MB, Harris NL (1990) Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol 183:252–278
Zhang XH, Chasin LA (2004) Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev 18(11):1241–1250. doi:10.1101/gad.1195304
Graveley BR (2000) Sorting out the complexity of SR protein functions. RNA 6(9):1197–1211
Manley JL, Krainer AR (2010) A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev 24(11):1073–1074. doi:10.1101/gad.1934910
Blencowe BJ, Bowman JA, McCracken S, Rosonina E (1999) SR-related proteins and the processing of messenger RNA precursors. Biochem Cell Biol 77(4):277–291
Long JC, Caceres JF (2009) The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417(1):15–27. doi:10.1042/BJ20081501
Fu XD (1995) The superfamily of arginine/serine-rich splicing factors. RNA 1(7):663–680
Zhu J, Mayeda A, Krainer AR (2001) Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol Cell 8(6):1351–1361
Tange TO, Damgaard CK, Guth S, Valcarcel J, Kjems J (2001) The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J 20(20):5748–5758. doi:10.1093/emboj/20.20.5748
Martinez-Contreras R, Fisette JF, Nasim FU, Madden R, Cordeau M, Chabot B (2006) Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol 4(2):e21. doi:10.1371/journal.pbio.0040021
Busch A, Hertel KJ (2012) Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA 3(1):1–12. doi:10.1002/wrna.100
Hoffman DW, Query CC, Golden BL, White SW, Keene JD (1991) RNA-binding domain of the A protein component of the U1 small nuclear ribonucleoprotein analyzed by NMR spectroscopy is structurally similar to ribosomal proteins. Proc Natl Acad Sci USA 88(6):2495–2499
Beyer AL, Christensen ME, Walker BW, LeStourgeon WM (1977) Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell 11(1):127–138
Han SP, Tang YH, Smith R (2010) Functional diversity of the hnRNPs: past, present and perspectives. Biochem J 430(3):379–392. doi:10.1042/BJ20100396
Mayeda A, Krainer AR (1992) Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68(2):365–375
Mayeda A, Munroe SH, Caceres JF, Krainer AR (1994) Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J 13(22):5483–5495
Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B (2007) hnRNP proteins and splicing control. Adv Exp Med Biol 623:123–147
Cho S, Moon H, Yang X, Zhou J, Kim HR, Shin MG et al (2012) Validation of trans-acting elements that promote exon 7 skipping of SMN2 in SMN2-GFP stable cell line. Biochem Biophys Res Commun 423(3):531–535. doi:10.1016/j.bbrc.2012.05.161
Goina E, Skoko N, Pagani F (2008) Binding of DAZAP1 and hnRNPA1/A2 to an exonic splicing silencer in a natural BRCA1 exon 18 mutant. Mol Cell Biol 28(11):3850–3860. doi:10.1128/MCB.02253-07
Tavanez JP, Madl T, Kooshapur H, Sattler M, Valcarcel J (2012) hnRNP A1 proofreads 3’ splice site recognition by U2AF. Mol Cell 45(3):314–329. doi:10.1016/j.molcel.2011.11.033
Bonnal S, Martinez C, Forch P, Bachi A, Wilm M, Valcarcel J (2008) RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol Cell 32(1):81–95. doi:10.1016/j.molcel.2008.08.008
Izquierdo JM, Majos N, Bonnal S, Martinez C, Castelo R, Guigo R et al (2005) Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell 19(4):475–484. doi:10.1016/j.molcel.2005.06.015
Izquierdo JM (2010) Cell-specific regulation of Fas exon 6 splicing mediated by Hu antigen R. Biochem Biophys Res Commun 402(2):324–328. doi:10.1016/j.bbrc.2010.10.025
Acknowledgments
We thank Juan Valcarcel for providing the mutant fas minigene plasmids (e1, E23, 6-6, 5-5). This work was supported by Mid-career Researcher Program through a National Research Foundation (NRF) grant (2011-0000188 and 20120005340) funded by the Ministry of Education, Science, and Technology (MEST), Korea; and a Systems Biology Infrastructure Establishment grant provided by Gwangju Institute of Science and Technology (GIST) in 2012.
Author information
Authors and Affiliations
Corresponding author
Additional information
Hyunkyung Oh and Eunkyung Lee have contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Oh, H.k., Lee, E., Jang, H.N. et al. hnRNP A1 contacts exon 5 to promote exon 6 inclusion of apoptotic Fas gene. Apoptosis 18, 825–835 (2013). https://doi.org/10.1007/s10495-013-0824-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-013-0824-8