Abstract
DNA analysis of blood meals from unfed nymphal Ixodes ricinus allows for the identification of tick host and tick-borne pathogens in the host species. The recognition of host species for tick larvae and the reservoirs of Borrelia, Rickettsia and Anaplasma species were simultaneously carried out by analysis of the blood meals of 880 questing nymphal I. ricinus ticks collected in forest parks of Szczecin city and rural forests in northwestern Poland that are endemic areas for Lyme borreliosis. The results obtained from the study indicate that I. ricinus larvae feed not only on small or medium animals but also on large animals and they (i.e. roe deer, red deer and wild boars) were the most prevalent in all study areas as the essential hosts for larvae of I. ricinus. The composition of medium and small vertebrates (carnivores, rodents, birds and lizards) provided a more diverse picture depending on study site. The reservoir species that contain the most pathogens are the European roe deer Capreolus capreolus, in which two species of Rickettsia and two species of Borrelia were identified, and Sus scrofa, in which one Rickettsia and three Borrelia species were identified. Rickettsia helvetica was the most common pathogen detected, and other included species were the B. burgdorferi s.l. group and B. miyamotoi related to relapsing fever group. Our results confirmed a general association of B. garinii with birds but also suggested that such associations may be less common in the transmission cycle in natural habitats than what was thought previously.
Similar content being viewed by others
Introduction
Ticks belonging to the Ixodidae family play an important role in the epidemiology of disease transmission to humans and animals. They are also an important element in maintaining the natural outbreaks of these diseases. Development and realisation of control procedures for tick-borne diseases require a detailed understanding of transmission pathways in the wild and, in particular, the identification of new reservoir hosts (Gariepy et al. 2012). Complete knowledge of the host-feeding patterns of tick populations in nature is a critical part of evaluating their vector competence and assessing the role of various vertebrates to serve as reservoir hosts of vector-borne pathogens (Kent 2010; Gariepy et al. 2012). The identification of tick hosts and reservoir hosts is difficult because it requires animal trapping and animal maintenance in the laboratory if xenodiagnosis is required. Host identification from blood meal sucked by a hematophagous vector has significantly facilitated such studies. DNA analysis of blood meal from unfed nymphal Ixodes ricinus allows identification of the hosts on which the larvae fed as well as tick-borne pathogens in the host species.
Molecular protocols for detection of host DNA are in constant development for the use of new targets and primers. For the differentiation of reservoirs and Borrelia species, Kirstein and Gray (1996) used the mitochondrially encoded cytb gene as the molecular target and digested the PCR products using the restriction endonucleases HaeIII and DdeI, a protocol that allowed to distinguish 11 animal species. Pichon et al. (2005) identified vertebrate DNA in the tick gut using PCR amplification with universal primers targeting a portion of the 18S rRNA gene, followed by reverse line blot (RLB) hybridisation. In the study of Humair et al. (2007), a 12S rDNA gene fragment was used for the first time as a PCR marker for detection of host DNA in nymphs.
The present paper is a study concerning molecular identification of I. ricinus hosts and Borrelia hosts by analysis of the blood meal in ticks in northwestern Poland using PCR–RFLP protocol with 12S rDNA gene fragment as molecular target. This research aims to identify host species of I. ricinus larvae, keeping in mind that immature stages of this tick species may feed not only on small animals but have the widest range of host species (L’Hostis et al. 1995; Kiffner et al. 2010). This research also aims to identify the reservoir species for Anaplasma phagocytophilum, Borrelia and Rickettsia and to elucidate any association between B. burgdorferi s.l. species and some vertebrate hosts by analysis of DNA from the midgut of blood-feeding nymphal ticks.
Materials and methods
Study material and collection sites of Ixodes ricinus
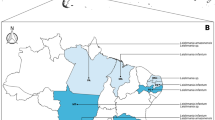
The study involved 880 specimens of unfed nymphs of I. ricinus collected in four study areas in northwestern Poland (Osow I, Szczecin Landscape Park II, Zielonczyn III and Pobierowo IV). These sites are known to be high risk areas for Borrelia infection in ticks (Wodecka 2003). Urban forests constitute 60 % of the city of Szczecin (northwestern Poland) and are mostly composed of forest complexes termed forest parks. The two selected sites, I and II, are the main recreational places for the population of Szczecin throughout the year. Site I is located in the city, Site II is on its outskirts, and large housing estates are situated within both areas. Site I is included in the forest complex of Szczecin, part of which lies at an altitude of 100–130 m above sea level, which provides a upland climate. The rest of the forests within Site I are located in the lowlands and depressions formed in areas rich in vegetation, peat meadows and marshes. In these biotopes, different groups of animals can be found, from invertebrates such as insects and arachnids, which are potential vectors for pathogens, to vertebrates, their potential reservoir. This is also a place for the convergence of bird flight paths in the meridional and latitudinal directions. One of the warmest climatic regions in Poland is Szczecin Landscape Park, which is influenced by the Atlantic climate and is characterised by small fluctuations in the daily and annual temperatures, mild winters, a high amount of rainfall per year and a good distribution of rainfall during the vegetation period. Both of these urban areas, Sites I and II, are characterised by different species of trees with a predominance of pines (Pinus spp.), oak (Quercus spp.), beech (Fagus spp.) and alder (Alnus spp.). Among the most common vegetation are fern (Polypodium vulgare), stinging nettle (Urtica dioica), mosses (Bryophyta spp.) and grass (Poaceae spp.). The other two tick collection sites were a mixture of forests often visited by strollers and mushroom pickers and farms (with a majority of Scots pine (Pinus silvestris), beech (Fagus sylvatica) and sessile oak (Quercus sessilis) with well-developed forest lining) located approximately 100 km to the west (Site III) and north (Site IV) of Szczecin. Zielonczyn, Site III, is composed of a small village around forest areas and wetlands, as well as pasture land in the Lower Oder Valley, which is located slightly further to the west. Water resources exist in the form of lakes and rivers located in the area, and a large forest area forms a temperate climate characterised by considerable humidity, which can be described as a warm temperate zone. Site IV includes forests near Pobierowo and sea bathing areas in northwestern Poland. The neighbouring marine basin, a pine forest with rich undergrowth, and vast meadows growing on peat contribute to the maintenance of this region-specific microclimate. Mild winters with a long frost-free period, high relative humidity and small temperature fluctuations throughout the year (IMWM 2010) favour the development of wildlife, especially small invertebrates that are involved in the circulation of microorganisms in the environment. Urban and rural forests are the habitat of many species of vertebrates, including large (wild boar, roe deer, red deer), medium and small (fox, raccoon, badger, marten, skunk, muskrat, rabbits, voles, shrew) animals. All sites have been monitored for ticks and tick-borne pathogens by us for several years (Skotarczak et al. 1999, 2008; Rymaszewska 2003; Wodecka 2003; Wodecka and Skotarczak 2005; Wodecka et al. 2010). Ticks were collected by sweeping up the vegetation up to 1 m with a flannel flag and then stored at −20 °C until DNA isolation.
DNA extraction
DNA extraction from ticks was carried out according to the phenol–chloroform protocol. All tick individuals (nymph or adult) were crushed using a ceramic pestle and suspended in 500 μl of 2× buffer (0.19 M NH4Cl, 0.011 M KHCO3 and 0.024 M EDTA) with the addition of 100 μl of Lysis buffer (0.017 M SDS, 0.01 M TRIS, 0.01 M EDTA) and 1 μl of Proteinase K (20 mg/ml) (BioShop, Canada). Subsequently, ticks were placed in a 56 °C water bath for 3 h. Following the incubation, 300 μl of phenol (BioShop) was added, and the tube was vortexed for 30 s and centrifuged for 10 min at 9,000 rpm. The supernatant was transferred successively to three additional tubes containing 400 μl of phenol–chloroform (1:1) and 300 μl of chloroform (POCH, Poland) (twice), then vortexed for 30 s and centrifuged for 10 min at 9,000 rpm. Finally, the supernatant was transferred to the last tube, and DNA was precipitated by adding 500 μl of isopropanol (POCH). The pellet was rinsed with 70 % ethanol and air-dried before suspension in Tris–EDTA (TE) buffer (pH 8.0) and the samples were stored at −70 °C until PCR analysis.
Detection of Ixodes ricinus host DNA in blood meal remnants by nested PCR
The mitochondrion-encoded 12S rDNA gene was used as the molecular marker for the detection of tick hosts by nested PCR. Two primer sets were used: outer 532fl2s (5′-CAAACTGGGATTAGATAC-3′) and 1l02rl2 s (5′-TGCTTACCTTGTTACGAC-3′), with a product length of approximately 520 bp, and inner 539f12s (5′-GGATTAGATACCCCACTATGC-3′) and 1015rl2 s (5′-TGAGGAGGGTGACGGGCGGT-3′), with a product length of approximately 440 bp.
The first PCR amplification was performed in a reaction volume of 10 μl containing 0.5 U of Allegro Taq DNA polymerase (Novazym, Poland), 70 mM Tris–HCl (pH 8.6 w 25 °C), 16.6 mM (NH4)2SO4, 3.5 mM MgCl2, 0.75 μM of deoxyribonucleoside triphosphate (Novazym), 2 pmol of the two outer primers, 532fl2s and 1l02rl2s, and 1 μl of the supernatant of the processed DNA sample in TE buffer. For the second PCR amplification, 1 μl of a 1:10 dilution of the first PCR product was added to 9 μl of reaction mixture prepared with the inner primers 539f12s and 102r12s. PCR was performed in thermal cyclers T-gradient (Biometria, Germany) and Peltier Thermal Cycler 200 (MJ Research, USA). Templates were subjected to an initial denaturation step of 94 °C for 10 min., followed by 40 cycles consisting of 94 °C for 30 s, 60 °C for 45 s and 72 °C for 1 min. In each PCR run the DNA of eleven vertebrate species were used as positive controls, i.e. Cervus elaphus, Capreolus capreolus, Dama dama and Sus scrofa (hunted near Dobra Szczecińska, Zachodniopomorskie province, Skotarczak et al. 2008), Vulpes vulpes, Meles meles and Nyctereutes procyonoides (hunted in Wielkopolska National Park) and Coccothraustes coccothraustes, Sturnus vulgaris, Turdus merula and Turdus philomelos (captured in Wielkopolska National Park, Michalik et al. 2008). TE buffer was applied as a negative control. The PCR products were analysed on a 1.5 % agarose gel at 5 V/cm for 1 h. Nova 100 DNA Ladder (Novazym, Poland) was applied for evaluation of the obtained product size. The results of the PCR amplification were viewed under UV light and were archived using BioCapt software (Vilber Lourmat, France).
Identification of Ixodes ricinus hosts by PCR–RFLP
The DNA amplified with primer set 539fl2s and 1015r12s was digested with enzymes AluI, Cfr13I, Tru1I, HpyF31, TaiI and BsuRI. These enzymes were selected on the basis of computer analysis (DNAMAN) of the 12S rDNA gene sequence downloaded from GenBank that shown there is possibility to obtain RFLP patterns of 60 vertebrate species. Among them were 23 bird species, 5 reptile species and 32 mammalian species (mice, hares, rabbits, bats, hedgehogs, minks, dormice, roe deer, wild boars, red deer, fallow deer)—the species represented in sites selected for the collection of I. ricinus.
Detection of Borrelia burgdorferi s.l. DNA by nested PCR–RFLP
A nested PCR method with two primer sets (outer 132f and 905r and inner 220f and 823r) was used to detect the flaB gene fragment of B. burgdorferi s.l. described earlier (Wodecka et al. 2010; Wodecka 2011). DNA isolated from a reference strain of B. burgdorferi s.s. IRS (German Collection of Microorganisms and Cell Cultures—DSMZ, Germany) was used as a positive control and TE buffer was used as a negative control. The PCR products were separated on a 1.5 % agarose gel (Prona, Spain) with the addition of ethidium bromide (Sigma-Aldrich, Germany) at 5 V/cm for 1 h. The MW1444 molecular marker (Polgen, Poland) was applied for evaluation of the size of the obtained product. The results of the PCR were viewed under UV light and were archived using BioCapt software (Vilber Lourmat, France). The DNA of flaB gene fragments amplified with primer set 220f and 823r were digested with enzymes HpyF3I and Ecl136II (Fermentas, Lithuania) to obtain the RFLP patterns of different Borrelia species, as described elsewhere (Wodecka 2011). The digestion products were analysed on a 3 % agarose gel at 5 V/cm for 2 h and archived as described above.
Detection of Anaplasma phagocytophilum and Rickettsia sp. DNA by PCR
The presence of A. phagocytophilum was detected with a 334 bp-long fragment of the msp2 gene marked by primers msp2-3F and msp2-3R (Levin et al. 2002). To detect the presence of Rickettsia DNA, we used the sequences of the gltA gene (citrate synthase), amplified using primers RpCS877 and RpCS1258 (382 bp product) (Nilsson et al. 1999). Phusion High-Fidelity DNA Polymerase (Finnzymes, Finland) was used for the PCR amplification at a mixture concentration 0.5 U/20 μl. Final reagent concentrations were 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM for each deoxynucleoside triphosphate, 10 pM for each primer and 2 μl DNA. The PCR regime was adjusted to the requirements of Phusion High-Fidelity DNA Polymerase according to the manufacturer’s recommendations. The results of PCR amplification were visualised by electrophoresis of 5–8 μl of each sample in 1.5 % agarose gels with ethidium bromide.
Contamination control procedures
To minimize contamination, the reagent setup, extraction, sample addition, and the PCR and sample analyses, were performed in 3 separate laboratories. Additionally, validation of restriction patterns for nested PCR products was performed by sequencing the amplicons obtained with primers 539f12s and 102r12s. Sequencing was performed for each positive control sample and study sample being representative of each restriction pattern for different vertebrate species. The 12S rRNA sequences of 32 strains determined in this study were deposited in GenBank under the accession numbers listed as follows: KF781309 (Accipiter gentilis), KF781310 and KF781334 (Capreolus capreolus), KF781311 (Castor fiber), KF781312 (Ciconia nigra), KF781313 and KF781335 (Cervus elaphus), KF781314 and KF781336 (Dama dama), KF781315 (Lacerta viridis), KF781316 (Lepus europaeus), KF781317 (Myodes glareolus), KF781318 and KF781338 (Meles meles), KF781319 (Natrix natrix), KF781320 and KF781321 (Oryctolagus cuniculus), KF781322 (Perdix perdix), KF781323 and KF781337 (Sus scrofa), KF781324, KF781325 and KF781330 (Turdus merula), KF781326 and KF781331 (Turdus philomelos), KF781327 (Upupa epops), KF781328 and KF781339 (Vulpes vulpes), KF781329 (Zootoca vivipara), KF781332 (Coccothraustes coccothraustes), KF781333 (Sturnus vulgaris), KF781340 (Nyctereutes procyonoides).
Results
Hosts DNA
DNA of potential I. ricinus larvae hosts were detected in 553 out of 880 nymphs (62.8 %). We obtained 19 types of restriction patterns consistent with the analysis of the 12S rRNA gene sequence. In total, 19 species of animals were identified: 10 mammals, 6 birds and 3 reptiles (Table 1). In each collection site the main hosts for I. ricinus larvae were large mammals, including wild boars, red deer and roe deer (Table 1). They constituted from 54.8 to 60 % of samples with detected host DNA and differences between every pair of compared sites were not statistically significant (p > 0.624). The differences between the same identified species were statistically significant in case of red deer when two forest parks were compared (p = 0.001) and in case of wild boars between each compared pair of sites (p < 0.049) excluding two rural forests comparison. The second most prevalent group of tick hosts in forest parks were birds and their participation ranged from 34.1 to 35.2 % of samples with detected host DNA in contrary to rural forests (5.5 and 6.8 %, Table 1). The differences were statistically significant when both types of habitats were compared (p < 0.001). Similar results were obtained in case of medium and small mammals that were the second most prevalent group of detected tick hosts in rural forest. In mentioned habitat they were identified in 29.5 % (Pobierowo) and in 30.8 % (Zielonczyn) whereas in forest parks their prevalence ranged from 4.8 to 7.5 % (Table 1). The differences were statistically significant when different habitats were compared (p < 0.001).
Pathogens’ DNA
DNA of tick-borne pathogens was detected using nested PCR protocol in 97 of 880 nymphs (11 %). The vast majority of pathogens detected in the nymphs were Rickettsia species (71.1 %, Table 2). The DNA of A. phagocytophilum was not detected in any ticks, but the DNA of Borrelia was affirmed in 27.8 % of all infected nymphs. B. garini and B. afzelii from the B. burgdorferi s.l. group were more frequent than B. miyamotoi of the relapsing fever group. Double infection of B. garinii and R. helvetica was observed once (Table 2).
Correlation of pathogens and hosts
The records of pathogen occurrence with respect to host DNA demonstrated that R. helvetica was mainly associated with Capreolus capreolus, Sus scrofa and Ce. elaphus, but the blood meal origin could not be determined in 37 ticks infected with this species (Table 2). DNA of B. afzelii was found in ticks that had fed on five species of animals, including large mammals (red deer, roe deer and wild boar) and medium ones (badger and rabbit). B. garinii also correlated with five host species, and B. miyamotoi with three host species (Table 2). The reservoir species that contained the most pathogens was the European roe deer C. capreolus, in which two species of Rickettcia and two species of Borrelia were identified: one from the B. burgdorferi s.l. group (B. afzelii) and one from the relapsing fever group, B. miyamotoi.
Discussion
The protocol applied in our study (PCR–RFLP) allows to detect 60 vertebrate species, including 32 mammalian species, 23 bird species, and five reptile species that are strictly connected with forest habitat and therefore may serve as hosts for ticks, especially for common tick I. ricinus. The DNA of potential hosts of I. ricinus larvae was detected in 553 nymphs (62.8 %). We obtained 19 types of restriction patterns consistent with those predicted on the basis of 12S rRNA gene sequences derived from GenBank.
Ticks were collected across a range of habitat types: two sites were chosen in the city and two in the rural forests. In each site the main hosts for I. ricinus larvae constitute large mammals (54.8–60 %) including roe deer, red deer and wild boars. Despite of accordance in the participation level of large mammals as tick hosts individual mammal species demonstrated site specific distribution: red deer were most prevalent in one forest park (Szczecin Landscape Park) and wild boars in the second (Osów) whereas in rural forests there were equal distributions of three mentioned species.
In the forest biotope of Szczecin (I-II), the crucial hosts for I. ricinus larvae, apart from large mammals, were birds. Sites I and II are frequently visited by humans, who are attacked but not constitute as hosts for I. ricinus larvae, nymphs and adults to maintain their population. Sites I and II are often exploited by dog owners, walkers, joggers and cyclists, which constrain undergrowth (mostly grasses, mosses, bilberries and ferns), such that only small game may inhabit these sites. In this case human rather disturb the vegetation scaring away small vertebrate fauna which may explain the lack of host rodents for larvae. Our study revealed that in these sites, this gap is filled by birds. In the contrary, the results of Estrada-Pena et al. (2005) showed a strong infestation of birds with the simultaneous presence of rodents in Spain. Pichon et al. (2005) found that songbirds are the most significant hosts in a forest on the outskirts of Berlin (Germany). In both above studies the results were obtained for the gene target located in the nucleus and for this reason the method would have discriminated in favour of birds comparing to our results.
In the two selected rural forest sites (III-IV), large mammals are also major hosts, though medium and small mammals and lizards also play significant roles. The results obtained in this and other studies suggest that the assumption that the immature stages of I. ricinus feed on small- and medium-sized mammals and that imago feed on large mammals (Kurtenbach et al. 1995; Liz et al. 2000; Hanincová et al. 2003a; Rizzoli et al. 2004; Michalik et al. 2005) should be verified. Thus, all stages of I. ricinus feed on roe deer (Kiffner et al. 2010), red deer and wild boars. It is known that the abundance of I. ricinus ticks is affected by habitat structure, climate and host community composition, but the claims by Jackson et al. (2006), Paziewska et al. (2010) and others—that changes in land use and greater human penetration of forest habitats are likely to have a greater force on the occurrence of tick-borne disease than climate changes—are well founded.
In the present study, the DNA of pathogens was detected in 97 out of 880 nymphs (11 %). The DNA of Borrelia was in 27.8 % of all infected nymphs and corresponded to 3 species of B. burgdorferi s.l. and one species of relapsing fever related B. miyamotoi. A double infection involving B. garinii and R. helvetica was observed once. Data on the host specificity of B. burgdorferi s.l. species (Gray et al. 2000; Kurtenbach et al. 2002; Hanincová et al. 2003b) suggest that B. valaisiana and B. garinii are mostly associated with birds, and the high proportion of these species at these sites points to birds as the main reservoir hosts of the species complex. The DNA analysis of tick hosts detected in blood meal remnants of I. ricinus nymphs in several contexts by Wodecka (2008) (Turdus merula, T. philomelos and Phasianus colchicus) and in the present study (Perdix perdix and Accipiter gentiles) confirms the conclusion above. Nevertheless, the DNA of B. garinii was detected alongside the DNA of Sus scrofa, Meles meles and Pedrix pedrix as well. The DNA of B. afzelii was found in ticks that had fed on 5 species of large and medium mammals, even though according to several authors, this species of spirochete is associated with rodents. Our results and those of other authors (Estrada-Pena et al. 2005; Pichon et al. 2006; Franke et al. 2010) seem to suggest that loose associations exist in the transmission cycle of Borrelia species in nature in various geographical areas. The DNA of B. miyamotoi was found in ticks that had fed on 4 species of animal hosts, including large mammals and hawks. B. lusitanie DNA was detected in the Polish population of I. ricinus for the first time in 2005 (Wodecka and Skotarczak 2005), and in the current study, DNA of host for this species was not identified.
The animal species in which the most Borrelia species occurred was Sus scrofa. The most pathogens were found in the European roe deer C. capreolus, in which two species of the Borrelia genus were identified, one from the B. burgdorferi s.l. group (B. afzelii) and one species of relapsing fever B. miyamotoi, as well as two species of Rickettsia.
Based on an analysis of unfed I. ricinus nymphs, Estrada-Pena et al. (2005) found that wild boar may be a reservoir species for B. afzelii. However, most of the evidence, especially that obtained from field studies, indicates a lack of reservoir competence of ungulate animals (European Union Concerted Action on Lyme Borreliosis—EUCALB). Generally, the ungulate animals play a large role in the epidemiology of borreliosis and the circulation of spirochetes in nature due to their role as hosts to I. ricinus ticks, which can be infected with pathogens through co-feeding with other ticks (Ogden et al. 1997; Juricová and Hubálek 2009). Thus far, it has been found that large forest mammals, such as wild boars, roe deer, red deer and those that are basic hosts for adult I. ricinus, are not able to constitute a reservoir for the species belonging to B. burgdorferi s.l. complex. Currently, many factors that determine a host’s sensitivity or resistance to Borrelia are known, but the crucial factor that could explain the selective transmission and host association of B. burgdorferi s.l. is the lytic component in serum, which was identified as the alternative pathway of complement (Kurtenbach et al. 2002). Analyses of resistance or sensitivity patterns to complement have been extended to many B. burgdorferi s.l. strains and different vertebrate species.
Perkins et al. (2006) demonstrated that animals such as deer are important blood hosts for feeding I. ricinus ticks but that they do not support transmission of many tick-borne pathogens, instead acting as dead-end transmission hosts. Deer maintain high tick intensities, which perpetuate tick populations, but they do not support tick-borne pathogen transmission, so they are dilution hosts. In our earlier molecular studies (Skotarczak et al. 2008) elucidating the role of game in the circulation of pathogens transmitted by ticks in northwestern Poland, DNA was isolated from tissues (blood and spleen) of roe deer, red deer and wild boars. The results showed that C. capreolus and Ce. elaphus play an important role as reservoirs of A. phagocytophilum, two Bartonella species, Babesia divergens and Theileria sp. but not that of Borrelia. Whereas the DNA of only one pathogen, A. phagocytophilum, occurred in the isolates obtained from 50 representatives of S. scrofa, in the present study, the DNA of A. phagocytophilum was not detected in any tick. However, that result is not surprising, as the presence of this pathogen is maintained at a relatively low level in north-western Poland (Rymaszewska 2004; Skotarczak et al. 2006, 2008).
The highest percentage of pathogens detected in the nymphs was the Rickettsia species (72 %). The results on pathogen occurrence in relation to DNA in blood meal demonstrated that R. helvetica was mainly associated with C. capreolus, S. scrofa and Ce. elaphus, but the blood meal origin could not be determined in 37 ticks infected with this bacterial species. R. helvetica is very widespread in Europe and has been detected in I. ricinus in several countries, including Italy (Beninati et al. 2002), Spain (Fernandez-Soto et al. 2004), Southern Germany (Hartelt et al. 2004) and Denmark and Sweden (Nilsson et al. 1999; Nielsen et al. 2004). The second species whose DNA we found together with roe deer DNA in engorged nymphal I. ricinus was R. monacensis, which is very rare in tick collection sites in north-western Poland (Rymaszewska and Piotrowski 2013) but is more often detected in I. ricinus in other countries, e.g. Germany (Schorn et al. 2011). R. monacensis was identified as an etiologic agent of a Mediterranean spotted fever (MSF)-like illness in Spain (Jado et al. 2007). Madeddu et al. (2012) reported a case of MSF-like illness in a 28-year-old man from Sardinia. I. ricinus ticks are considered to be vectors of R. monacensis and have been found in Sardinia, although less often than other tick species. However, other ticks might act as vectors for R. monacensis in Sardinia, where ticks of the genus Rhipicephalus are prominent. The identification of R. monacensis as a cause of an MSF-like illness expands the list of pathogenic rickettsiae circulating in Italy.
Known species of Rickettsia are the only tick-borne pathogens transmitted transovarially with 100 % efficiency. Generally, tick-transmitted Rickettsia species persist for a short time in vertebrate organisms, making the bacteremia difficult to detect (Raoult and Roux 1997). Nevertheless, recent studies indicate that some vertebrate animals may be a reservoir for Rickettsia. De Sousa et al. (2012) indicated a potential role of the Teira dugesii lizard species in the maintenance and transmission cycle of I. ricinus tick-borne agents such as R. monacensis and R. helvetica that are circulating on Madeira Island, and Franke et al. (2010) considered the reservoir role of birds on a conservation island in the Baltic Sea. Additionally, our study (unpublished data) revealed the presence of R. helvetica DNA in the blood of a goat Capra hircus (9 %). In Japan, two independent teams confirmed the presence of R. helvetica DNA in the blood of sika deer (Cervus nippon yesoensis) and in raccoons (Procyon lotor) (7.1 and 1.6 %, respectively) (Inokuma et al. 2008; Sashika et al. 2010). In addition, Seino et al. (2008) detected a related species, R. asiatica, in the blood of C. nippon yesoensis in Hokkaido.
The presented research results show that the nymphal I. ricinus ticks, which parasitize deer in the larval stage, were most often infected by Rickettsia. A relatively high level of infection may indicate that the infection is not coincidental and that these results are not simply a consequence of the transovarial transfer of rickettsiae. Therefore, the hypothesis about the role of ruminants in the circulation of bacteria of the genus Rickettsia seems probable and should be confirmed in further studies.
References
Beninati T, Lo N, Noda H, Esposito F (2002) First detection of spotted fever group rickettsiae in Ixodes ricinus from Italy. Emerg Infect Dis 8:983–986
De Sousa R, Lopes de Carvalho I, Santos AS, Bernardes C, Milhano N, Jesus J, Menezes D, Nuncio MS (2012) Role of the lizard Teira dugesii as a potential host for Ixodes ricinus tick-borne pathogens. Appl Environ Microbiol 78(10):3767–3769
Estrada-Pena A, Osacar JJ, Pichon B, Gray JS (2005) Host and pathogen detection for immature stages of Ixodes ricinus (Acari: Ixodidae) in north-central Spain. Exp Appl Acarol 37:257–268
Fernandez-Soto P, Perez-Sanchez R, Encinas-Grandes A, Sanz RA (2004) Detection and identification of Rickettsia helvetica and Rickettsia sp. IRS3/IRS4 in Ixodes ricinus ticks found on humans in Spain. Eur J Clin Microbiol Infect Dis 23:648–649
Franke J, Meier F, Moldenhauer A, Straube E, Dorn W, Hildebrandt A (2010) Established and emerging pathogens in Ixodes ricinus ticks collected from birds on a conservation island in the Baltic Sea. Med Vet Entomol 24(4):425–432
Gariepy TD, Lindsay R, Ogden N, Gregory TR (2012) Identifying the last supper: utility of the DNA barcode library for bloodmeal identification in ticks. Mol Ecol Resour 12:646–652
Gray JS, Robertson JN, Key S (2000) Limited role of rodents as reservoirs of Borrelia burgdorferi sensu lato in Ireland. Eur J Epidemiol 16:101–103
Hanincová K, Schäfer SM, Etti S, Sewell HS, Taragelova V, Ziak D, Labuda M, Kurtenbach K (2003a) Association of Borrelia afzelii with rodents in Europe. Parasitology 126(Pt 1):11–20
Hanincová K, Taragelová V, Koci J, Schäfer SM, Hails R, Ullmann AJ, Piesman J, Labuda M, Kurtenbach K (2003b) Association of Borrelia garinii and B. valaisiana with songbirds in Slovakia. Appl Environ Microbiol 69:2825–2830
Hartelt K, Oehme R, Frank H, Brockmann SO (2004) Pathogens and symbionts in ticks: prevalence of Anaplasma phagocytophilum (Ehrlichia sp.), Wolbachia sp., Rickettsia sp., and Babesia sp. in Southern Germany. Int J Med Microbiol 293:86–92
Humair PF, Douet V, Morán Cadenas F, Schouls LM, Van De Pol I, Gern L (2007) Molecular identification of bloodmeal source in Ixodes ricinus ticks using 12S rDNA as a genetic marker. J Med Entomol 44:869–880
Inokuma H, Seino N, Suzuki M, Kaji K, Takahashi H, Igota H, Inoue S (2008) Detection of Rickettsia helvetica DNA from peripheral blood of sika deer (Cervus nippon yesoensis) in Japan. J Wild Dis 44:164–167
Institute of Meteorology and Water Menagement—National Research Institute of Poland (2010) Monthly Climate Monitoring Bulletin 2010
Jackson LE, Hilborn ED, Thomas JC (2006) Towards landscape design guidelines for reducing Lyme disease risk. Int J Epidemiol 35:315–322
Jado I, Oteo JA, Aldámiz M, Gil H, Escudero R, Ibarra V, Portu J, Portillo A, Lezaun MJ, García-Amil C, Rodríguez-Moreno I, Anda P (2007) Rickettsia monacensis and human disease, Spain. Emerg Infect Dis 13(9):1405–1407
Juricová Z, Hubálek Z (2009) Serologic survey of the wild boar (Sus scrofa) for Borrelia burgdorferi sensu lato. Vec Bor Zoon Dis 9(5):479–482
Kent RJ (2010) Molecular methods for arthropod bloodmeal identification and applications to ecological and vector-borne disease studies. Mol Ecol Resour 9:4–18
Kiffner C, Lödige C, Alings M, Vor T, Rühe F (2010) Abundance estimation of Ixodes ticks (Acari: Ixodidae) on roe deer (Capreolus capreolus). Exp Appl Acarol 52(1):73–84
Kirstein F, Gray JS (1996) A molecular marker for the identification of the zoonotic reservoirs of Lyme borreliosis by analysis of the blood meal in its European vector, Ixodes ricinus. Appl Environ Microbiol 62:4060–4065
Kurtenbach K, Kampen H, Dizij A, Arndt S, Seitz HM, Schaible UE, Simon MM (1995) Infestation of rodents with larval Ixodes ricinus (Acari: Ixodidae) is an important factor in the transmission cycle of Borrelia burgdorferi s.l. in German woodlands. J Med Entomol 32(6):807–817
Kurtenbach K, De Michelis S, Etti S, Schäfer SM, Sewell HS, Brade V, Kraiczy P (2002) Host association of Borrelia burgdorferi sensu lato—the key role of host complement. Trends Microbiol 10(2):74–79
Levin ML, Nicholson WL, Massung RF, Sumner JW, Fish D (2002) Comparison of the reservoir competence of medium-sized mammals and Peromyscus leucopus for Anaplasma phagocytophilum in Connecticut. Vec Bor Zoon Dis 2(3):125–136
L’Hostis M, Dumon H, Dorchies B, Boisdron F, Gorenflot A (1995) Seasonal incidence and ecology of the tick Ixodes ricinus (Acari: Ixodidae) on grazing pastures in Western France. Exp Appl Acarol 19:211–220
Liz JS, Anderes L, Sumner JW, Massung RF, Gern L, Rutti B, Brossard M (2000) PCR detection of granulocytic ehrlichiae in Ixodes ricinus ticks and wild small mammals in Western Switzerland. J Clin Microbiol 38(3):1002–1007
Madeddu G, Mancini F, Caddeo A, Ciervo A, Babudieri S, Maida I, Fiori ML, Rezza G, Mura MS (2012) Rickettsia monacensis as cause of Mediterranean spotted fever–like illness, Italy. Emerg Infect Dis 18(4):702–704
Michalik J, Skotarczak B, Skoracki M, Wodecka B, Sikora B, Hofman T, Rymaszewska A, Sawczuk M (2005) Borrelia burgdorferi sensu stricto in yellow-necked mice and feeding Ixodes ricinus ticks in a forest habitat of west central Poland. J Med Entomol 5:850–856
Michalik J, Wodecka B, Skoracki M, Sikora B, Stańczak J (2008) Prevalence of avian-associated Borrelia burgdorferi s.l. genospecies in Ixodes ricinus ticks collected from blackbirds (Turdus merula) and song thrushes (T. philomelos). Int J Med Microbiol 298(S1):129–138
Nielsen H, Fournier PE, Pedersen IS, Krarup H (2004) Serological and molecular evidence of Rickettsia helvetica in Denmark. Scand J Infect Dis 36:559–563
Nilsson K, Lindquist O, Liu AJ, Jaenson TG, Friman G, Pahlson C (1999) Rickettsia helvetica in Ixodes ricinus ticks in Sweden. J Clin Microbiol 37(2):400–403
Ogden NH, Nuttall PA, Randolph SE (1997) Natural Lyme disease cycles maintained via sheep by co-feding ticks. Parasitology 115:591–599
Paziewska A, Zwolińska L, Harris PD, Bajer A, Siński E (2010) Utilization of rodent species by larvae and nymphs of hard ticks (Ixodidae) in two habitats in NE Poland. Exp Appl Acarol 50:79–91
Perkins SE, Cattadori IM, Tagliapietra V, Rizzoli AP, Hudson PJ (2006) Localized deer absence leads to tick amplification. Ecology 87(8):1981–1986
Pichon B, Rogers M, Egan D, Gray J (2005) Blood-meal analysis for the identification of reservoir hosts of tick-borne pathogens in Ireland. Vec Bor Zoon Dis 5:172–180
Pichon B, Kahl O, Hammer B, Gray JS (2006) Pathogens and host DNA in Ixodes ricinus nymphal ticks from German forest. Vec Bor Zoon Dis 6:382–387
Raoult D, Roux V (1997) Rickettsioses as paradigms of new or emerging infectious diseases. Clin Microbiol Rev 10:694–719
Rizzoli A, Rosà R, Mantelli B, Pecchioli E, Hauffe H, Tagliapietra V, Beninati T, Neteler M, Genchi C (2004) Ixodes ricinus, transmitted diseases and reservoirs (article in Italian). Parassitologia 46(1–2):119–122
Rymaszewska A (2003) Detection of human factor granulocytic ehrlichiosis in ticks Ixodes ricinus in Western Pomerania. In: Buczek A, Błaszak C (eds) Arthropods and hosts. LIBER Publishing House, Lublin, pp 175–184
Rymaszewska A (2004) Comparing the sensitivity of Anaplasma phagocytophilum DNA detection in Ixodes ricinus ticks by amplifying a fragment of the epank-1 and 16S rDNA genes. Folia Med Cracov 3–4:79–85
Rymaszewska A, Piotrowski J (2013) Use of DNA sequences for Rickettsia identification in Ixodes ricinus ticks: the first detection of Rickettsia monacensis in Poland. Microbes Infect 15:140–146
Sashika M, Abe G, Matsumoto K, Inokuma H (2010) Molecular survey of rickettsial agents in feral raccoons (Procyon lotor) in Hokkaido, Japan. Jpn J Infest Dis 63:353–354
Schorn S, Pfister K, Reulen H, Mahling M, Silaghi C (2011) Occurrence of Babesia spp., Rickettsia spp. and Bartonella spp. in Ixodes ricinus in Bavarian public parks, Germany. Parasit Vec 4:135
Seino JN, Matsumoto K, Hayakawa D, Suzuki M, Hata H, Kondo S, Yokoyama N, Inokuma H (2008) Serological and molecular survey of rickettsial infection in cattle and sike deer in a pastureland in Hidaka district, Hokkaido, Japan. Jpn J Infect Dis 61:315–317
Skotarczak B, Soroka M, Wodecka B (1999) The occurrence of Ixodes ricinus in the selected recreational areas in the Province of Szczecin. Part I. Wiad Parazytol 45:507–517
Skotarczak B, Rymaszewska A, Wodecka B, Sawczuk M, Adamska M, Maciejewska A (2006) PCR detection of granulocytic Anaplasma and Babesia in Ixodes ricinus ticks and birds in west-central Poland. Ann Agric Environ Med 13:21–23
Skotarczak B, Adamska M, Sawczuk M, Maciejewska A, Wodecka B, Rymaszewska A (2008) Coexistence of tick-borne pathogens in game animals and ticks in western Poland. Vet Med 53(12):668–675
Wodecka B (2003) Detection of DNA of Borrelia burgdorferi sensu lato in Ixodes ricinus ticks in north-western Poland. Ann Agric Environ Med 10:171–178
Wodecka B (2011) FlaB gene as a molecular marker for distinct identification of Borrelia species in environmental samples by the PCR-restriction fragment length polymorphism method. Appl Environ Microbiol 77:7088–7092
Wodecka B, Skotarczak B (2005) First isolation of Borrelia lusitaniae DNA from Ixodes ricinus ticks in Poland. Scan J Infect Dis 37:27–34
Wodecka B, Leońska A, Skotarczak B (2010) A comparative analysis of molecular markers for the detection and identification of Borrelia spirochetes in Ixodes ricinus. J Med Microbiol 59:309–314
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Wodecka, B., Rymaszewska, A. & Skotarczak, B. Host and pathogen DNA identification in blood meals of nymphal Ixodes ricinus ticks from forest parks and rural forests of Poland. Exp Appl Acarol 62, 543–555 (2014). https://doi.org/10.1007/s10493-013-9763-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10493-013-9763-x