Abstract
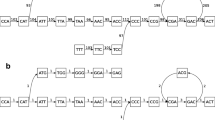
DNA reassembling is an NP-hard problem (Brun, Theor Comput Sci 395:31–46, 2008; Medvedev et al 2007; Ma and Lombardi 2008). The present article presents a locally guided global learning system to solve the problem of genome reassembling. We have used a reference DNA sequence which is 99 % similar to an unknown DNA sequence. Two different sequences from the same organism generally have around 99 % similarity (Wei et al 2007). We have considered different DNA sequences from NCBI website (http://www.ncbi.nlm.nih.gov). Then we have simulated the tasks of cloning the sequence, followed by shearing the clones to a number of short reads. In our algorithm, we have introduced a new concept in the task of DNA reassembling using Ant Colony Optimization, where pheromone concentration is proportional to the score of assembled DNA fragments with some known reference sequences within the same organism. Unlike local overlapping, we have used here local alignment score of short reads with some known local reference region as the heuristic information. The result shows that our algorithm is capable of reassembling at par with the state-of-the-art. DNA reassembling techniques may need a massive parallel computation and huge memory space (Kurniawan et al 2008) because of size ~109bp of DNA sequences of mammals (Miller et al, Genomics 95:315–327, 2010; Blazewicz et al, Comput Biol Chem 33:224–230, 2009; Butler et al, Genome Res 18:810–820, 2008; Joshi et al 2011; Stupar et al, Arch Oncol 19:3–4, 2011; Quail et al, BMC Genomics 13:1471–2164, 2012), and ACO is inherently concurrent in nature (Dorigo and Stutzle 2004). Due to lack of appropriate computational resources, we had to confine ourselves to deal with the sequences of length up to ∼105 b p. We have considered 22 sequences of different organism, including Homo sapiens BRCA1 (127429bp) gene. For large sequences, we have applied hierarchical BAC-by-BAC sequencing (Fig. 2) (Myers, Comput Sci Eng 1:33–43, 1999), to stitch the individual segments to retrieve the original DNA sequence.




Similar content being viewed by others
References
Garca S, Molina D, Lozano M, Herrera F (2009) A study on the use of non-parametric tests for analyzing the evolutionary algorithms behaviour: a case study on the CEC2005 Special Session on Real Parameter Optimization. Journal of Heuristcs 15:617– 644
Indumathy R, Uma Maheswari S (2014) Solving DNA Sequence Assembly Using Particle Swarm Optimization With Inertia Weight and Constriction Factor. International Journal of Soft Computing and Artificial Intelligence 2(1):90–94
Verma RS, Singh V, Kumar S (2011) DNA Sequence Assembly using Particle Swarm Optimization. Int J Comput Appl 28(10):34–38
Fang S-C, Wang Y, Zhong J (2005) A Genetic Algorithm Approach to Solving DNA Fragment Assembly Problem. J Comput Theor Nanosci 2:1–7
Parsons RJ, Forrest S, Burks C (1995) Genetic algorithms, operators, and DNA fragment assembly. Mach Learn 21(1-2):11–33
Nebro AJ, Luque G, Luna F, Alba E (2008) DNA fragment assembly using a grid-based genetic algorithm. Comput Oper Res 35(9):2776–2790
Luque G, Alba E, Khuri S (2005) Parallel Computing for Bioinformatics and Computational Biology, WILEY, Chapter-12: Assembling DNA Fragments with a Distributed Genetic Algorithm
Karaboga D, Akay B (2009) A comparative study of Artificial Bee Colony algorithm. Appl Math Comput 25:108–132
Karaboga D, Ozturk C, Karaboga N, Gorkemli B (2012) Artificial bee colony programming for symbolic regression. Inf Sci 209:01–15
Firoz JS, Sohel Rahman M, Saha TK (2012) Bee Algorithms for Solving DNA Fragment Assembly Problem with Noisy and Noiseless data. GECCO ’12 Proceedings 14th Annual Conference on Genetic and Evolutionary Computation. ACM, NY, pp 201–208
Ansorge WJ (2009) Next generation DNA sequencing techniques. New Biotechnol 25(4):167–260
Blazewicz J, Bryjaa M, Figlerowicz M, Gawrona P, Kasprzak M, Kirton E, Platt D, Przybytek J, Swiercz A, Szajkowski L (2009) Whole genome assembly from 454 sequencing output via modified DNA graph concept. Comput Biol Chem 33:224–230
Blum C, Valles MY, Blesa MJ (2008) An ant colony optimization algorithm for DNA sequencing by hybridization. Comput Oper Res 35:362–3635
Brun Y (2008) Solving NP-complete problems in the tile assembly model. Theor Comput Sci 395:31–46
Butler J, MacCallum I, Kleber M, Shlyakhter IA, Belmonte MK, Lander ES, Nusbaum C, Jaffe DB (2008) Allpaths: De novo assembly of whole-genome shotgun micro reads. Genome Res 18:810–820
Dorigo M, Maniezzo V, Colorni A (1996) Ant system: optimization by a colony of cooperating agents. IEEE Trans Systems Man Cybern Part B 26:29–41
Dorigo M, Stutzle T (2004) Ant Colony Optimization. MIT Press, London
Isakov O, Shomron N Deep sequencing data analysis: Challenges and solutions. Bioinformatics Trends and Methodologies, Intech, November 2011, ch-29:Deep Sequencing Data Analysis
Joshi N, Srivastava S, Kumar M, Kavalan J, Karandikar SK, Saraph A (2011) Parallelization of velvet, a de-novo genome sequence assembler. IEEE International Conference on High Performance Computing
Kurniawan TB, Ibrahim Z, Saaid MFM, Yahya A (2008) Implementation of ant system for DNA sequence optimization. NANO-SciTech, Shah Alam
Ma X, Lombardi F (2008) Combinatorial optimization problem in designing DNA self-assembly tile sets. 2008 IEEE International Workshop on Design and Test of Nano Devices, Circuits and Systems, pp 73–76
Medvedev P, Georgiou K, Myers G, Brudno M (2007) Computability models of sequence assembly. Workshop on Algorithms in Bioinformatics, Philadelphia, 289–301
Meksangsouy P, Chaiyaratana N (2003) DNA fragment assembly using an ant colony system algorithm. Proceedings Evolutionary Computation. CEC ’03 3:1756–1763
Miller JR, Koren S, Sutton G (2010) Assembly algorithms for next generation sequencing data. Genomics 95:315–327
Myers G (1999) Whole-genome dna sequencing. Comput Sci Eng 1:33–43
Myllykangas S, Buenrostro J, Ji HP (2012) Overview of sequencing technology platforms. Bioinformatics for High Throughput Sequencing, 11–25
Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Yong G (2012) A tale of three next generation sequencing platforms: comparison of ion torrent, pacific biosciences and illumina Miseq sequencers. BMC Genom 13:1471–2164
Scheibye-Alsing K, Hoffmann S, Frankel AM, Jensen P, Stadler PF (2009) Sequence assembly. Comput Biol Che:33
Stupar M, Vidovi V, Luka D (2011) Functions of human non-coding DNA sequences. Arch Oncol 19:3–4
Treangen TJ, Salzberg SL (2011) Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet 113(1):36–46
Wei L-T, Yang C-B, Ann H-Y, Peng Y-H (2007) Ant colony optimization algorithms for sequence assembly with haplotyping. 6th Conference on Information Technology and Applications in Outlying Islands, Yunlin, Taiwan, 260–268
Zerbino DR, Velvet EB (2008) Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 1:821–829
Fullwood MJ, Wei C-L, Liu ET (2009) Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res 19:521–532
Author information
Authors and Affiliations
Corresponding author
Additional information
Rights and permissions
About this article
Cite this article
Baidya, S., De, R.K. A novel locally guided genome reassembling technique using an artificial ant system. Appl Intell 43, 397–411 (2015). https://doi.org/10.1007/s10489-015-0650-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-015-0650-5