Abstract
A polyphasic study was undertaken to determine the taxonomic status of a Streptomyces strain which had been isolated from a high altitude Atacama Desert soil and shown to have bioactive properties. The strain, isolate H9T, was found to have chemotaxonomic, cultural and morphological properties that place it in the genus Streptomyces. 16S rRNA gene sequence analyses showed that the isolate forms a distinct branch at the periphery of a well-delineated subclade in the Streptomyces 16S rRNA gene tree together with the type strains of Streptomyces crystallinus, Streptomyces melanogenes and Streptomyces noboritoensis. Multi-locus sequence analysis (MLSA) based on five house-keeping gene alleles showed that isolate H9T is closely related to the latter two type strains and to Streptomyces polyantibioticus NRRL B-24448T. The isolate was distinguished readily from the type strains of S. melanogenes, S. noboritoensis and S. polyantibioticus using a combination of phenotypic properties. Consequently, the isolate is considered to represent a new species of Streptomyces for which the name Streptomyces aridus sp. nov. is proposed; the type strain is H9T (=NCIMB 14965T=NRRL B65268T). In addition, the MLSA and phenotypic data show that the S. melanogenes and S. noboritoensis type strains belong to a single species, it is proposed that S. melanogenes be recognised as a heterotypic synonym of S. noboritoensis for which an emended description is given.
Similar content being viewed by others
Introduction
Streptomycetes remain a unique source of novel pharmaceutically important products with antibacterial, anti-inflammatory and antitumor activities (Bérdy 2005; Demain 2014; Barka et al. 2016), hence the continued interest in these organisms as a source of new specialised metabolites (Goodfellow and Fiedler 2010; Chaudhary et al. 2013). However, the continued search for new bioactive compounds against drug-resistant microorganisms needs to be focused on novel streptomycetes to avoid the rediscovery of known compounds from common Streptomyces species. Given this imperative, novel Streptomyces species are being sought from unusual and neglected habitats (Hong et al. 2009; Tiwari and Gupta 2012), notably from extreme biomes (Bull 2011; Hamedi et al. 2013; Goodfellow 2013). Previous work from our group has shown that novel Streptomyces species abound in arid Atacama Desert soils (Okoro et al. 2009; Busarakam 2014; Busarakam et al. 2014), some of which synthesise new specialised metabolites with encouraging bioactivities (Bull et al. 2016), thereby underpinning the premise that extreme environmental conditions promote unique actinobacterial diversity which is the basis of novel chemistry (Bull and Stach 2007; Bull 2011; Gomez-Escribano et al. 2015).
Streptomyces, the type genus of the family Streptomycetaceae, was proposed by Waksman and Henrici (1943) and the description of the taxon emended by Witt and Stackebrandt (1990) and Wellington et al. (1992). The genus encompasses aerobic, Gram-stain positive actinobacteria with a high DNA G+C content, which form extensively branched substrate mycelia supporting aerial hyphae that typically differentiate into chains of spores, have a wall peptidoglycan rich in LL-diaminopimelic acid, contain major amounts of saturated, iso- and anteiso- fatty acids, usually have either hexa- or octa-hydrogenated menaquinones with nine isoprene units as predominant isoprenologues and complex polar lipid patterns which tend to include diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol and phosphatidylinositol mannosides (Kämpfer 2012). The genus includes over 700 validly named species (http://www.bacterio.net/streptomyces.html) which can be assigned to many multi- and single-membered subclades in the Streptomyces 16S rRNA gene tree (Kämpfer 2012; Labeda et al. 2012) and based on multi-locus sequence analysis (MLSA; Labeda et al. 2017). Despite being the largest genus in the domain Bacteria the taxon remains underspeciated (Okoro et al. 2009; Antony-Babu et al. 2010; Busarakam 2014). New species are assigned to the genus using a combination of genotype and phenotype properties, and some recent studies have featured MLSA (Rong and Huang 2012, 2014; Labeda et al. 2014, 2017; Labeda 2016).
The aim of the present study was to establish the taxonomic position of a Streptomyces strain, isolate H9T, which had been isolated from a high altitude Atacama Desert soil and shown to produce novel specialised metabolites (Idris 2016). The results of a polyphasic taxonomic study showed that isolate H9T belongs to a novel Streptomyces species for which we propose the name Streptomyces aridus sp. nov.
Materials and methods
Selective isolation, maintenance and cultural conditions
Strain H9T was recovered from an arid subsurface soil sample (30 cm depth) at 4000 metres above sea level on Cerro Chajnantor (23o63′31″S, 67o52′27″W) east of San Pedro de Atacama, Chile. The strain was isolated on glucose-yeast extract agar (Athalye et al. 1981) supplemented with cycloheximide and nystatin (each at 25 µg/ml) after incubation at 28 °C for 14 days following inoculation with a suspension of one gram of soil in 5 ml ¼ strength Ringer’s solution. The isolate together with S. melanogenes NRRL B-2072T, S. noboritoensis NRRL B-12152T and S. polyantibioticus NRRL B-24448T were maintained on yeast extract-malt extract agar (International Streptomyces Project [ISP medium 2], Shirling and Gottlieb 1966). All of the reference strains were obtained from the NRRL culture collection as indicated.
Biomass for the chemotaxonomic and molecular systematic analyses was prepared in shake flasks (180 revolutions per minute) of ISP2 broth following incubation at 28 °C for 14 days and washed twice in distilled water; cells for the chemotaxonomic studies were freeze-dried and those for the molecular systematic analyses stored at room temperature.
Chemotaxonomy
Strain H9T was examined for chemotaxonomic properties known to be characteristic of Streptomyces strains (Kämpfer 2012). Standard procedures were used to detect isomers of diaminopimelic acid (A2pm) (Staneck and Roberts 1974), menaquinones (Collins et al. 1985), polar lipids (Minnikin et al. 1984) and whole cell sugar composition (Lechevalier and Lechevalier 1970), using appropriate controls. The type strains of S. melanogenes and S. noboritoensis were also included in the analyses for A2pm isomers, cellular sugars and polar lipids. Fatty acid methyl esters (FAMEs) were prepared from isolate H9T and the type strains of S. melanogenes, S. noboritoensis and polyantibioticus by saponification, methylation and extraction following protocols developed by Miller (1982) with minor modifications from Kuykendall et al. (1988). The FAMEs were separated by gas chromatography (Agilent 6890 N instrument) and the resultant peaks automatically integrated. Fatty acid names and properties were determined using the standard microbial identification (MIDI) system Version 4.5 and the ACTIN 6 database (Sasser 1990).
Phylogenetic analyses
16S rRNA gene sequencing
Genomic DNA was extracted from isolate H9T biomass and PCR-mediated amplification of a 16S rRNA purified gene product obtained, as described by Kim and Goodfellow (2002). Identification of phylogenetic neighbours and calculation of pairwise 16S rRNA gene sequence similarities were realised using the EzTaxon-e server (http://eztaxon-e.ezbiocloud.net/; Kim et al. 2012). The CLUSTAL W algorithm from MEGA 6 (Tamura et al. 2013) was used to align the sequences. Phylogenetic trees were generated using the maximum-likelihood, maximum-parsimony and neighbour-joining algorithms drawn from the MEGA 6 software package. Evolutionary distances were calculated using the Kimura two-parameter (Kimura 1980) and the topologies of the resultant trees evaluated by bootstrap analyses (Felsenstein 1985) based on 1000 replicates. The trees were rooted using the 16S rRNA gene sequence of Streptomyces albus subsp. albus DSM 40313T (GenBank accession number AJ 621602).
Multi-locus sequence analyses
The experimental and data handling procedures used in the MLSA were based upon modifications of described procedures (Labeda et al. 2014, 2017; Labeda 2016). Genomic DNA was isolated from the strain using an UltraClean® Microbial DNA isolation kit (MoBio Labs, Carlsbad, CA) by following the manufacturer’s instructions. Partial sequences of the house-keeping genes atpD (ATP synthase F1, beta subunit), gyrB (DNA gyrase B subunit), rpoB (RNA polymerase beta subunit), recA (recombinase A) and trpB (tryptophan synthetase, beta subunit) were amplified and sequenced using primers and protocols, as described previously by Labeda et al. 2014. The amplified products were purified using ExoSAP-IT (Affymetrix, Santa Clara, CA), sequenced using BigDye 3.1 on an ABI sequencer model 3730 and assembled using Sequencher version 5.2 (Gene Codes, Ann Arbor, MI).
The gene sequences for the 5 house-keeping loci of the strain were deposited in GenBank (see Supplemental Table S1) and were also organised using the Bacterial Isolate Genomic Sequence Database (BIGSdb) version 1.12.3 (Jolley and Maiden 2010) on the ARS Microbial Genomic Sequence Database server (http://199.133.98.43). Genome sequences, where available, were uploaded into the sequence bin for the respective isolates in the BIGSdb isolate database. The genome sequences were scanned within BIGSdb for house-keeping loci, the sequences of which were then tagged and the allele sequences and respective allele designations added to the sequence database when new alleles were found. The strain record was then updated with the matching allele identification for each locus held in the strain database. The sequences for the alleles of the loci of isolate H9T were individually aligned with MAFFT (Katoh and Standley 2013), subsequently concatenated head to tail in-frame, and exported in FASTA format, providing a dataset of 706 Streptomyces strains and 2622 positions (Labeda 2016).
Phylogenetic relationships were constructed in IQ-Tree version 1.41 (Nguyen et al. 2015) using the maximum-likelihood algorithm based on the general time reversible model (Nei and Kumar 2000 with invariable sites plus a discrete Gamma-model based on 4 rate categories (Gu et al. 1995) which had been shown to be the optimal model for such data using iModelTest 2 (Darriba et al. 2012). The individual trees were the subject of 1000 ultrafast bootstrap replications (Minh et al. 2013) followed by 1000 replications of assessment of branch supports with single branch tests using the SH-like approximate likelihood ration test (Guindon et al. 2010). MLSA evolutionary distances were determined using MEGA 6 by calculating the Kimura 2-parameter distances (Kimura 1980). Strain pairs having ≤0.007 MLSA evolutionary distances were considered conspecific based on the guideline empirically determined by Rong and Huang (2012), namely that this MLSA distance (Kimura 2-parameter distance), computed from the partial sequences of these house-keeping loci, equates to the 70% DNA:DNA cut-off point recommended for the delineation of prokaryotic species by Wayne et al. (1987).
Cultural and morphological properties
The cultural features of isolate H9T and the type strains of S. melanogenes, S. noboritoensis and S. polyantibioticus were recorded on tryptone-yeast extract, yeast extract-malt extract, oatmeal, inorganic salts-starch, glycerol-asparagine, peptone-yeast extract-iron and tyrosine agar plates (ISP media 1–7, Shirling and Gottlieb 1966) that had been incubated for 14 days at 28 °C. Spore chain morphology and spore surface ornamentation were detected following growth on oatmeal agar (ISP medium 3; Shirling and Gottlieb 1966) for 14 days at 28 °C, by scanning electron microscopy (Cambridge 240 instrument) after O’Donnell et al. (1993).
Phenotypic tests
Strain H9T and the type strains of the three reference Streptomyces species were examined for a range of standard biochemical, degradative and physiological properties using media and methods described by Williams et al. (1983). Enzyme profiles of the strains were determined using API ZYM kits (bioMérieux) following the manufacturer’s instructions. A standard inoculum corresponding to 5 on the McFarland scale (Murray et al. 1999) was used to inoculate all of these tests. In addition, the ability of the strains to oxidise diverse carbon and nitrogen sources and to show resistance to inhibitory compounds were determined using GEN III microplates in an Omnilog device (BIOLOG Inc., Haywood, USA). The exported data were analysed using the opm package for R (Vaas et al. 2012, 2013) version 1.06. All of these tests were carried out in duplicate.
Results and discussion
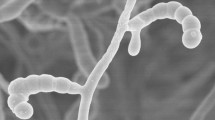
The chemotaxonomic, cultural and morphological properties of strain H9T were found to be consistent with its classification in the genus Streptomyces (Kämpfer 2012). The organism forms an extensively branched substrate mycelium which carries aerial hyphae that differentiate into spiral chains of smooth surfaced spores on all of the ISP media tested, as exemplified in Fig. 1. The strain forms brown to black substrate mycelia on most of the ISP media and either mild brown or brown black diffusible pigments on ISP media 5–7 (Table 1). Whole organism hydrolysates of the strain were found to be rich in LL-A2pm, glucose, mannose and ribose; the predominant fatty acids were identified as anteiso-C15:0 (34.6%), iso-C16:0 (19.4%), anteiso-C17:0 (17.9%) and C16:0 (10.9%) (Table 2); the major isoprenologues were identified as tetra-, hexa- and octa-hydrogenated menaquinones (16, 23 and 30%, respectively); and the polar lipid pattern was found to consist of diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol, glycophosphatidylinositol, one unidentified lipid and an aminolipid (Fig. S1).
Isolate H9T was found to form a distinct branch at the periphery of a well delineated subclade in the Streptomyces 16S rRNA gene tree together with S. crystallinus NBRC 15401T, S. melanogenes NBRC 12890T and S. noboritoensis NRRL B-12152T, a relationship that was supported by all of the tree-making algorithms but not by a high bootstrap value (Fig. 2). The isolate is closely related to the S. melanogenes and S. noboritoensis strains sharing a 16S rRNA gene sequence similarity with them of 98.6%, a value found to correspond to 20 nucleotide (nt) differences at 1424 and 1423 locations respectively; the S. melanogenes and S. noboritoensis strains were shown to have identical 16S rRNA gene sequences. A subclade consisting of S. melanogenes NBRC 12890T and S. noboritoensis NRRL B-12152T was identified as cluster 30 in the 16S rRNA gene analysis of Labeda et al. (2012), S. polyantibioticus NRRL B-24448 T was found adjacent to this taxon. The corresponding 16S rRNA gene sequence homologies between the isolate and the remaining phylogenetically close strains were found to fall within the range 97.7–98.5%, values shown to equate to 21 and 31 nt differences.
Neighbour-joining phylogenetic tree based on nearly complete 16S rRNA gene sequences (1329–1425 nucleotides) showing relationships between isolate H9T and closely related type strains of Streptomyces species. Asterisks indicate branches of the tree that were recovered using the maximum-likelihood (ML) and maximum-parsimony tree-making methods. ML indicates branches of the tree that were also supported by this algorithm. Numbers at the modes indicate levels of bootstrap support based on a neighbour-joining analysis of 1000 resampled datasets, only values above 50% are given. The scale bar indicates 0.005 substitutions per nucleotide position
MLSA have been found to clarify relationships between closely related streptomycetes because of the strong phylogenetic signal provided by partial sequences of single-copy house-keeping genes (Rong and Huang 2012, 2014; Labeda 2011, 2016, Labeda et al. 2014, 2016, 2017); In the present MLSA analysis, the relationships found between isolate H9T and the type strains of closely related Streptomyces species are shown in Fig. 3 and Table 3. The isolate is closely related to S. melanogenes NRRL B-2072T, S. noboritoensis NRRL B-1252T and S. polyantibioticus NRRL B-24448T, relationships that are supported by a 100% bootstrap value; S. crystallinus NRRL B-3629T is loosely associated with this lineage. The relationship of S. crystallinus NBRC 15401T, S. melanogenes NBRC 12890T and S. noboritoensis NRRL B-12152T was also identified in a recent MLSA study (Labeda et al. 2017). Isolate H9T was shown to have MLSA distances greater than 0.007 with all of these strains indicating that it forms the nucleus of a novel Streptomyces species. In contrast, the type strains of S. melanogenes and S. noboritoensis share a MLSA evolutionary distance of only 0.004 indicating that they belong to the same genomic species.
Streptomyces sub-tree derived from the phylogenetic tree inferred from concatenated partial sequences of the house-keeping genes atpD, gyrB, recA, rpoB and trpB using the maximum-likelihood method based on the General Time Reversible model. The final dataset consisted of 2622 positions and 706 strains. Percentages at the nodes represent levels of bootstrap support from 1000 resampled datasets, values less than 60% are not shown. The proposed new species is indicated in bold. Bar, equals number of substitutions per site
Identical results were obtained for nearly all of the duplicated phenotypic tests, the exceptions being a few carbon source tests recorded from the GEN111 microplates. It can be seen from Table 4 that the isolate can be distinguished from the type strains of S. melanogenes, S. noboritoensis and S. polyantibioticus, its close phylogenetic neighbours, by a broad range of phenotypic tests though all four strains have many properties in common. In particular, the isolate can be distinguished from the three reference type strains by its ability to form spiral chains of spores, grow at 40 °C and oxidise d-fucose and d-raffinose. Conversely, the three reference type strains can be differentiated from isolate H9T by their ability to form straight to flexuous spore chains and degrade arbutin, hypoxanthine and l-tyrosine. In turn, the isolate can be distinguished from S. melanogenes NRRL B-2072T and S. noboritoensis NRRL B-12151T by its ability to degrade casein, starch and Tween 80, but not arbutin, guanine or urea.
It is apparent that the type strains of S. melanogenes and S. noboritoensis have many phenotypic properties in common, notably morphological and cultural features found to be of particular value in the circumscription of Streptomyces species by Labeda et al. (2012) in their phylogenetic survey of Streptomycetaceae species. The present results are in good agreement with those of previous studies in which the type strains of S. melanogenes and S. noboritoensis were assigned to the same numerically defined phenotypic clusters (Williams et al. 1983; Kämpfer et al. 1991) and the same MLSA lineage (Labeda et al. 2017). In addition, all three strains were shown to have whole organism hydrolysates rich in LL-A2pm, glucose, mannose and ribose and similar polar lipid patterns (Fig S1). Similar chemotaxonomic markers have been recorded for the type strain of S. polyantibioticus, NRRL B-2443T (Roes and Meyers 2009). The fatty acid profiles of the type strains of S. melanogenes, S. noboritoensis and S. polyantibioticus, like that of isolate H9T, were shown to contain major proportions of anteiso-C15:0, iso-C16:0, C16:0 and anteiso-C17:0 though quantitative differences were apparent; such differences were also detected between other fatty acids though a few trace components were discontinuously distributed (Table 2).
In summary, isolate H9T is only loosely associated with its near phylogenetic neighbours in the Streptomyces 16S rRNA gene tree and its distinctness is strongly supported by corresponding MLSA data based on concatenated sequences of five house-keeping genes. It can also be distinguished from the type strains of S. melanogenes and S. noboritoensis, its close phylogenetic neighbours, based on a combination of phenotypic properties. These data clearly show that isolate H9T forms a new centre of taxonomic variation within the genus Streptomyces. The name proposed for this taxon is Streptomyces aridus sp. nov. It is also clear from the MLSA and associated chemotaxonomic and phenotypic data that the type strains of S. melanogenes and S. noboritoensis belong to the same species. It is, therefore, proposed that S. melanogenes Suguwara and Onuma (1957) be seen as a heterotypic synonym of S. noboritoensis Isono et al. (1957). An emended description is given of the latter.
Description of Streptomyces aridus sp. nov.
Streptomyces aridus (a’ri. dus. L. masc. adj. aridus, dry, referring to the isolation of the strain from arid soil).
Aerobic, Gram-stain positive, catalase positive actinobacterium that forms an extensively branched substrate mycelium that bears aerial hyphae that differentiate into spiral chains of smooth surfaced spores (1–1.5 µm × 0.5 µm) on oatmeal agar. A brown–black diffusible pigment is produced on yeast extract-malt extract agar. Grows from 10 to 40 °C, optimally ~28 °C, from pH 5–10, optimally ~pH 7.0 and in the presence of up to 2.5% w/v NaCl. Additional cultural and phenotypic features are cited in the text and in Tables 1 and 4. Chemotaxonomic characteristics are typical of the genus Streptomyces. The type strain, H9T (=NCIMB 14965T=NRRL B-65268T), was isolated from a subsurface soil sample collected at 4000 metres above sea level on Cerro Chajnantor, east of San Pedro de Atacama in north eastern Chile. The GenBank accession number for the 16S rRNA gene sequence of isolate H9T is LT594571.
Emended description of Streptomyces noboritoensis Isono et al. 1957, 21AL
Heterotypic synonym: Streptomyces melanogenes Suguwara and Onuma 1957, 141AL.
Data taken from the present study and from Kämpfer (2012).
Aerobic, Gram-stain positive actinobacterium which forms extensively branched substrate mycelia that bear aerial hyphae which differentiate into straight to filamentous spore chains. Mature spore chains are long with 10–50, or often more than 50, spores per chain. This morphology is seen on glycerol-asparagine agar, oatmeal agar, salts-starch agar and yeast extract-malt extract agar. Spore surface is smooth. Grows at 10, 20 and 30 °C but not at 4 or 40 °C, at pH6 and pH7 and in the presence of 4%, w/v sodium chloride. Additional cultural and phenotypic properties are cited in the text and in Tables 1 and 4. Chemotaxonomic features are typical of the genus Streptomyces.
The type strain of S. noboritoensis (NRRL B-12152T), was isolated from soil from Inada-noborito, Kawasaki City, Kanagawa Prefecture, Japan.
Type strain: ATCC 23937, CBS 92168, DSM 40192, NBRC 12390, JCM 4378, NCIMB 9835, NRRL B-2072, RIA 1146.
References
Antony-Babu S, Stach JEM, Goodfellow M (2010) Computer-assisted numerical analysis of colour-group data for dereplication of streptomycetes for bioprospecting and ecological purposes. Antonie Van Leeuwenhoek 97:231–239. doi:10.1007/s10482-009-9404-x
Athalye M, Lacey J, Goodfellow M (1981) Selective isolation and enumeration of actinomycetes using rifampicin. J Appl Bacteriol 51:289–297. doi:10.1111/j.1365-2672.1981.tb01244.x
Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, Klenk HP, Clément C, Ouhdouch Y, van Wezel GP (2016) Taxonomy, physiology, and natural products of actinobacteria. Microbiol Mol Biol Rev 80:1–43. doi:10.1128/mmbr.00019-15
Bérdy J (2005) Bioactive microbial metabolites. J Antibiot 58:1–26
Bull AT (2011) Actinobacteria of the extremobiosphere. In: Horikoshi K (ed) Extremophiles Handbook. Springer, Tokyo, pp 1203–1240
Bull AT, Stach JEM (2007) Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol 15:491–499. doi:10.1016/j.tim.2007.10.004
Bull AT, Asenjo JA, Goodfellow M, Gómez-Silva B (2016) The Atacama Desert: technical resources and the growing importance of novel microbial diversity. Ann Rev Microbiol 70:215–234. doi:10.1146/annurev-micro-102215-095236
Busarakam K (2014) Novel actinobacterial diversity in arid Atacama Desert soils as a source of new drug leads. PhD thesis, Newcastle University, UK
Busarakam K, Bull AT, Girard G, Labeda DP, van Wezel GP, Goodfellow M (2014) Streptomyces leeuwenhoekii sp. nov., the producer of chaxalactins and chaxamycins, forms a distinct branch in Streptomyces gene trees. Antonie Van Leeuwenhoek 105:849–861. doi:10.1007/s10482-014-0139-y
Chaudhary HS, Soni B, Shrivastava AR, Shrivastava S (2013) Diversity and versatility of actinomycetes and its role in antibiotic production. J Appl PharmSc 3:S83–S94. doi:10.7324/JAPS.2013.38.S14
Collins MD, Goodfellow M, Minnikin DE, Alderson G (1985) Menaquinone composition of mycolic acid-containing actinomycetes and some sporoactinomycetes. J Appl Bacteriol 58:77–86. doi:10.1111/j.1365-2672.1985.tb01431.x
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and high-performance computing. Nat Methods 9:772. doi:10.1038/nmeth.2109
Demain A (2014) Importance of microbial natural products and the need to revitalize their discovery. J Ind Microbiol Biotechnol 41:185–201. doi:10.1007/s10295-013-1325-z
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gomez-Escribano JP, Castro JF, Razmilic V, Chandra G, Andrews BA, Asenjo JA, Bibb MJ (2015) The Streptomyces leeuwenhoekii genome: de novo sequencing and assembly in single contigs of the chromosome, circular plasmid pSLE1 and linear plasmid pSLE2. BMC Genom 16:1–11. doi:10.1186/s12864-015-1652-8
Goodfellow M (2013) Actinobacterial diversity as a sources of new drugs. Microbiologist 14:8–12
Goodfellow M, Fiedler HP (2010) A guide to successful bioprospecting: informed by actinobacterial systematics. Antonie Van Leeuwenhoek 98:119–142. doi:10.1007/s10482-010-9460-2
Gu X, Fu YX, Li WH (1995) Maximum likelihood estimation of the heterogeneity of substitution rate among nucleotide sites. Mol Biol Evol 12:546–557
Guindon S, Dufayard J, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi:10.1093/sysbio/syq010
Hamedi J, Mohammadipanah F, Ventosa A (2013) Systematic and biotechnological aspects of halophilic and halotolerant actinomycetes. Extremophiles 17:1–13. doi:10.1007/s00792-012-0493-5
Hong K, Gao A, Xie Q, Gao H, Zhuang L, Lin H, Yu H, Li J, Yao X, Goodfellow M, Ruan J (2009) Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar Drugs 7:24–44
Idris H (2016) Actinobacterial diversity in Atacama Desert habitats as a road map to biodiscovery. PhD thesis, Newcastle University, UK
Isono K, Tyamashita S, Tomiyama Y, Suzuki S, Sakai H (1957) Studies on homomycin II. J Antibiotics (Tokyo) Series A 10:21–30
Jolley KA, Maiden MCJ (2010) BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinform 11:1–11. doi:10.1186/1471-2105-11-595
Kämpfer P (2012) Genus Streptomyces. In: Goodfellow M, Kämpfer P, Busse HJ, Trujillo ME, Suzuki K-i, Ludwig W, Whitman WB (eds) Bergey’s manual of systematic bacteriology, vol 5. The Actinobacteria, vol 2. Springer, New York, pp 1455–1767
Kämpfer P, Kroppenstedt RM, Dott W (1991) A numerical classification of the genera Streptomyces and Streptoverticillium using miniaturized physiological tests. J Gen Microbiol 137:1831–1891
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi:10.1093/molbev/mst010
Kim SB, Goodfellow M (2002) Streptomyces thermospinisporus sp. nov., a moderately thermophilic carboxydotrophic streptomycete isolated from soil. Int J Syst Evol Microbiol 52:1225–1228. doi:10.1099/00207713-52-4-1225
Kim O, Cho Y, Lee K, Yoon S, Kim M, Na H, Park S, Jeon Y, Lee J, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721. doi:10.1099/ijs.0.038075-0
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kuykendall LD, Roy MA, O’Neill JJ, Devine TE (1988) Fatty acids, antibiotic resistance and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int J Syst Evol Microbiol 38:358–361. doi:10.1099/00207713-38-4-358
Labeda DP (2011) Multi-locus sequence analysis of phytopathogenic species of Streptomyces. Int J Syst Evol Microbiol 61:2525–2531
Labeda DP (2016) Taxonomic evaluation of putative Streptomyces scabiei strains held in the ARS culture collection (NRRL) using multi-locus sequence analysis. Antonie Van Leeuwenhoek 109:349–356. doi:10.1007/s10482-015-0637-6
Labeda DP, Goodfellow M, Brown R, Ward AC, Lanoot B, Vanncanneyt M, Swings J, Kim SB, Liu Z, Chun J, Tamura T, Oguchi A, Kikuchi T, Kikuchi H, Nishii T, Tsuji K, Yamaguchi Y, Tase A, Takahashi M, Sakane T, Suzuki KI, Hatano K (2012) Phylogenetic study of the species within the family Streptomycetaceae. Antonie Van Leeuwenhoek 101:73–104. doi:10.1007/s10482-011-9656-0
Labeda DP, Doroghazi JR, Ju KS, Metcalf WW (2014) Taxonomic evaluation of Streptomyces albus and related species using multilocus sequence analysis and proposals to emend the description of Streptomyces albus and describe Streptomyces pathocidini sp. nov. Int J Syst Evol Microbiol 64:894–900. doi:10.1099/ijs.0.058107-0
Labeda DP, Rong X, Huang Y, Doroghazi JR, Ju K-S, Metcalf WW (2016) Taxonomic evaluation of species of the Streptomyces hirsutus clade using multi-locus sequence analysis and proposals to reclarify several species in this clade. Int J Syst Evol Microbiol 66:2444–2450
Labeda DP, Dunlap CA, Rong X, Huang Y, Doroghazi JR, Ju K-S, Metcalf WW (2017) Phylogenetic relationships in the family Streptomy cetaceae using multi-locus sequence analysis Antonie van Leeuwenhoek (in press)
le Roes-Hill M, Meyers PR (2009) Streptomyces polyantibioticus sp. nov., isolated from the banks of a river. Int J Syst Evol Microbiol 59:1302–1309. doi:10.1099/ijs.0.006171-0
Lechevalier MP, Lechevalier HA (1970) Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Evol Microbiol 20:435–443. doi:10.1099/00207713-20-4-435
Miller LT (1982) Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J Clin Microbiol 16:584–586
Minh BQ, Nguyen MAT, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195. doi:10.1093/molbev/mst024
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Murray PR, Boron EJ, Pfaller MA, Tenover FC, Yolken RH (1999) Manual of Clinical Microbiology, 7th edn. ASM Press, Washington, DC
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
Nguyen L, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi:10.1093/molbev/msu300
O’Donnell AG, Falconer C, Goodfellow M, Ward AC, Williams E (1993) Biosystematics and diversity amongst novel carboxydotrophic actinomycetes. Antonie Van Leeuwenhoek 64:325–340. doi:10.1007/BF00873091
Okoro CK, Brown R, Jones AL, Andrews BA, Asenjo JA, Goodfellow M, Bull AT (2009) Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert, Chile. Antonie Van Leeuwenhoek 95:121–133. doi:10.1007/s10482-008-9295-2
Rong X, Huang Y (2012) Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA–DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst Appl Microbiol 35:7–18. doi:10.1016/j.syapm.2011.10.004
Rong X, Huang Y (2014) Multi-locus sequence analysis: taking prokaryotic systematics to the next level. Methods Microbiol 41:221–251. doi:10.1016/bs.mim.2014.10.001
Sasser MJ (1990) Identification of bacteria by gas chromatography of cellular fatty acids. Del: Microbial ID Inc, Newark
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Evol Microbiol 16:313–340
Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231
Suguwara R, Onuma M (1957) Melanomycin, a new antitumor substance from Streptomyces. II. Description of strain. J Antibiotics (Tokyo) 10:138–142
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tiwari K, Gupta RK (2012) Rare actinomycetes: a potential storehouse for novel antibiotics. Crit Rev Biotech 32:108–132. doi:10.3109/07388551.2011.562482
Vaas LAI, Sikorski J, Michael V, Göker M, Klenk HP (2012) Visualization and curve-parameter estimation strategies for efficient exploration of phenotype microarray kinetics. PLoS ONE 7:e34846. doi:10.1371/journal.pone.0034846
Vaas LAI, Sikorski J, Hofner B, Fiebig A, Buddruhs N, Klenk HP, Göker M (2013) opm: an R package for analysing OmniLog® phenotype microarray data. Bioinformatics. doi:10.1093/bioinformatics/btt291
Waksman SA, Henrici AT (1943) The nomenclature and classification of the actinomycetes. J Bacteriol 46:337–341
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Trüper HG (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464. doi:10.1099/00207713-37-4-463
Wellington EMH, Stackebrandt E, Sanders D, Wolstrup J, Jorgensen NO (1992) Taxonomic status of Kitasatospora, and proposed unification with Streptomyces on the basis of phenotypic and 16S rRNA analysis and emendation of Streptomyces Waksman and Henrici 1943, 339AL. Int J Syst Bacteriol 42:156–160
Williams ST, Goodfellow M, Alderson G, Wellington EMH, Sneath PHA, Sackin MJ (1983) Numerical classification of Streptomyces and related genera. J Gen Microbiol 129:1743–1813
Witt D, Stackebrandt E (1990) Unification of the genera Streptoverticillum and Streptomyces, and emendation of Streptomyces Waksman and Henrici 1943, 339AL. Syst Appl Microbiol 13:361–371
Acknowledgements
This project was partly funded by a UK Newton Project for UK-Chile collaboration (JIC CA 586). We thank the staff of the European Southern Observatory for permission and assistance in collecting soil samples from the Chajnantor Plateau. The able technical assistance of E. Basehoar in determining the house keeping gene sequences is gratefully acknowledged. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer. H.I. is grateful to the Malaysian Government for a Ph.D. scholarship and M.G. for an Emeritus Fellowship from the Leverhulme Trust. DPL and the ARS Culture Collection CRIS project was supported by ARS National Program 301.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Idris, H., Labeda, D.P., Nouioui, I. et al. Streptomyces aridus sp. nov., isolated from a high altitude Atacama Desert soil and emended description of Streptomyces noboritoensis Isono et al. 1957. Antonie van Leeuwenhoek 110, 705–717 (2017). https://doi.org/10.1007/s10482-017-0838-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-017-0838-2