Abstract
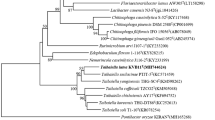
A Gram-stain negative, aerobic, rod-shaped and non-spore-forming bacterium, CC-Bw-6T was isolated from chopped tomato stems. Analysis of the 16S rRNA gene sequence of the strain CC-Bw-6T showed its affiliation with the genus Lysobacter within the class Gammaproteobacteria. Strain CC-Bw-6T was found to be most closely related to Lysobacter panaciterrae KCTC 12601T (97.0 %) and Lysobacter daecheongensis KCTC 12600T (96.8 %), and showed lower similarity (<96.5 %) to other Lysobacter species. DNA–DNA relatedness between strain CC-Bw-6T and L. panaciterrae KCTC 12601T was 10.8 %, the G+C content of the genomic DNA is 69.9 mol%. Strain CC-Bw-6T was determined to possess C11:0 iso, C11:0 iso 3OH, C14:0 iso, C15:0 anteiso, C15:1 ω5c, C16:1 ω5c, C16:0, C15:0 iso, C16:0 iso, C17:0 iso and C16:0 10-methyl/C17:1 iso ω9c as predominant fatty acids. The major polar lipid profile consists of phosphatidylethanolamine, phosphatidylglycerol, diphosphatidylglycerol and unidentified phospholipids. The predominant polyamine is spermidine, and ubiquinone-8 is the predominant respiratory quinone. Analysis of phenotypic, geno- and phylogenetic characteristics revealed a distinct taxonomic position of strain CC-Bw-6T with respect to other Lysobacter species. Based on the phylogenetic, chemotaxonomic and phenotypic data presented here, we propose a novel species with the name Lysobacter lycopersici sp. nov. The type strain is CC-Bw-6T (=BCRC 80612T = JCM 19164T).


Similar content being viewed by others
References
Ahmed K, Chohnan S, Ohashi H, Hirata T, Masaki T, Sakiyama F (2003) Purification, bacteriolytic activity, and specificity of β-lytic protease from Lysobacter sp. IB-9374. J Biosci Bioeng 95:27–34
Bae HS, Im WT, Lee ST (2005) Lysobacter concretionis sp. nov., isolated from anaerobic granules in an upflow anaerobic sludge blanket reactor. Int J Syst Evol Microbiol 55:1155–1161
Bernardet JF, Nakagawa Y, Holmes B (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070
Choi J-H, Seok J-H, Cha J-H, Cha C-J (2014) Lysobacter panacisoli sp. nov., isolated from ginseng soil. Int J Syst Evol Microbiol 64:2193–2197
Chou JH, Cho NT, Arun AB, Young CC, Chen WM (2008) Luteimonas aquatica sp. nov., isolated from fresh water from Southern Taiwan. Int J Syst Evol Microbiol 58:2051–2055
Christensen P, Cook FD (1978) Lysobacter, a new genus of nonfruiting, gliding bacteria with a high base ratio. Int J Syst Bacteriol 28:367–393
Collins MD (1985) Isoprenoid quinone analysis in classification and identification. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 267–287
Dittmer JC, Lester RL (1964) A simple, specific spray for the detection of phospholipids on thin-layer chromatograms. J Lipid Res 15:126–127
Edwards U, Rogall T, Blocker H, Emde M, Bottger EC (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17:7843–7853
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Finkmann W, Altendorf K, Stackebrandt E, Lipski A (2000) Characterization of N2O-producing Xanthomonas-like isolates from biofilters as Stenotrophomonas nitritireducens sp. nov., Luteimonasmephitis gen. nov., sp. nov. and Pseudoxanthomonas broegbernensis gen. nov., sp. nov. Int J Syst Evol Microbiol 50:273–282
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Folman LB, De Klein MJEM, Postma J, van Veen JA (2004) Production of antifungal compounds by Lysobacter enzymogenes isolate 3.1T8 under different conditions in relation to its efficacy as a biocontrol agent of Pythium aphanidermatum in cucumber. Biol Control 31:145–154
Graham PH, Sadowsky MJ, Keyser HH, Barnet YM, Bradley RS, Cooper JE, De Ley DJ, Jarvis BDW, Roslycky EB et al (1991) Proposed minimal standards for the description of new genera and species of root-and stem-nodulating bacteria. Int J Syst Bacteriol 41:582–587
Heiner CR, Hunkapiller LK, Chen SM, Glass JI, Chen EY (1998) Sequencing multimegabase-template DNA using BigDye terminator chemistry. Genome Res 8:557–561
Jung HM, Ten LN, Im WT, Yoo SA, Lee ST (2008) Lysobacter ginsengisoli sp. nov., a novel species isolated from soil in Pocheon Province, South Korea. J Microbiol Biotechnol 18:1496–1499
Kawamura Y, Tomida J, Morita Y, Naka T, Mizuno S, Fujiwara N (2009) ‘Lysobacter enzymogenes ssp. cookii’ Christensen 1978 should be recognized as an independent species, Lysobacter cookii sp. nov. FEMS Microbiol Lett 298:118–123
Kilic-Ekici O, Yuen GY (2003) Induced resistance as a mechanism of biological control by Lysobacter enzymogenes strain C3. Biol Control 93:1103–1110
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Mergaert J, Cnockaert MC, Swings J (2003) Thermomonas fusca sp. nov. and Thermomonas brevis sp. nov., two mesophilic species isolated from a denitrification reactor with poly(ε-caprolactone) plastic granules as fixed bed, and emended description of the genus Thermomonas. Int J Syst Evol Microbiol 53:1961–1966
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Miller LT (1982) Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxyl acids. J Clin Microbiol 16:584–586
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal K, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Murray RGE, Doetsch RN, Robinow CF (1994) Determination and cytological light microscopy. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington DC, pp 31–32
Ngo HTT, Won K, Du J, Son H-M, Park Y, MooChang K, Kim K-Y, Jin F-X, Yi T-H (2014) Lysobacter terrae sp. nov. isolated from Aglaia odorata rhizosphere soil. Int J Syst Evol Microbiol. doi:10.1099/ijs.0.067397-0
Oh KH, Kang SJ, Jung YT, Oh TK, Yoon JH (2011) Lysobacter dokdonensis sp. nov., isolated from soil. Int J Syst Evol Microbiol 61:1089–1093
Paisley R (1996) MIS whole cell fatty acid analysis by gas chromatography training manual. MIDI, Newark
Park JH, Kim R, Aslam Z, Jeon CO, Chung YR (2008) Lysobacter capsici sp. nov., with antimicrobial activity, isolated from the rhizosphere of pepper, and emended description of the genus Lysobacter. Int J Syst Evol Microbiol 58:387–392
Romanenko LA, Uchino M, Tanaka N, Frolova GM, Mikhailov VV (2008) Lysobacter spongiicola sp. nov., isolated from a deep-sea sponge. Int J Syst Evol Microbiol 58:370–374
Romanenko LA, Tanaka N, Svetashev VI, Kurilenlo VV, Mikhailov VV (2013) Luteimonas vadosa sp. nov., isolated from seashore sediment. Int J Syst Evol Microbiol 63:1261–1266
Ryazanova LP, Stepnaya OA, Suzina NE, Kulaev IS (2005) Antifungal action of the lytic enzyme complex from Lysobacter sp. XL1. Process Biochem 40:557–564
Saddler GS, Bradbury JF (2005) Family I. Xanthomonadaceae fam. nov. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM (eds) Bergey’s manual of systematic bacteriology (The Proteobacteria), part B (The Gammaproteobacteria). Springer, New York, p 63
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc, Newark, DE
Scherer P, Kneifel H (1983) Distribution of polyamines in methanogenic bacteria. J Bacteriol 154:1315–1322
Seldin L, Dubnau D (1985) Deoxyribonucleic acid homology among Bacillus polymyxa, Bacillus macerans, Bacillus azotofixans, and other nitrogen-fixing Bacillus strains. Int Syst Bacteriol 35:151–154
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Ten LN, Jung H-M, Im W-T, Yoo S-A, Lee S-T (2008) Lysobacter daecheongensis sp. nov., isolated from sediment of stream near the Daechung Dam in south Korea. J Microbiol 46:519–524
Ten LN, Jung H-M, Im W-T, Yoo S-A, Oh HM, Lee S-T (2009) Lysobacter panaciterrae sp. nov., isolated from soil of a ginseng field. Int J Syst Evol Microbiol 59:958–963
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE (1987) International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Weon HY, Kim BY, Kim MK, Yoo SH, Kwon SW, Go SJ, Stackebrandt E (2007) Lysobacter niabensis sp. nov. and Lysobacter niastensis sp. nov., isolated from greenhouse soils in Korea. Int J Syst Evol Microbiol 57:548–551
Worliczek HL, Kämpfer P, Rosengarten R, Tindall BJ, Busse H-J (2007) Polar lipid and fatty acid profiles-re-vitalizing old approaches as a modern tool for the classification of mycoplasmas? Syst Appl Microbiol 30:355–370
Yang S-Z, Feng G-D, Zhu H-H, Wang Y-H (2014) Lysobacter mobilis sp. nov., isolated from abandoned lead-zinc ore. Int J Syst Evol Microbiol. doi:10.1099/ijs.0.000026
Yassin AF, Chen W-M, Hupfer H, Siering C, Kroppenstedt RM, Arun AB, Lai W-A, Shen F-T, Rekha PD, Young C-C (2007) Lysobacter defluvii sp. nov., isolated from municipal solid waste. Int J Syst Evol Microbiol 57:1131–1136
Ye X-M, Chu C-W, Shi C, Zhu J-C, He Q, He J (2014) Lysobacter caeni sp. nov., isolated 1 from the sludge of pesticide manufacturing factory. Int J Syst Evol Microbiol. doi:10.1099/ijs.0.000026
Young C-C, Kämpfer P, Chen W-M, Yen W-S, Arun AB, Lai W-A, Shen F-T, Rekha PD, Lin K-Y, Chou J-H (2007) Luteimonas composti sp. nov., a moderately thermophilic bacterium isolated from food waste. Int J Syst Evol Microbiol 57:741–744
Zhang L, Bai J, Wang Y, Wu GL, DaiJ Fang C (2011) Lysobacter korlensis sp. nov. and Lysobacter bugurensis sp. nov., isolated from soil in north-west China. Int J Syst Evol Microbiol 61:2259–2265
Acknowledgments
Authors would like to thank Professor Dr. Hans G. Trüper for his advice regarding the nomenclature. We also thank Dr. Mariyam Shahina for technical assistance. This research work was kindly supported by grants from the National Science Council, the Council of Agriculture, Executive Yuan and in part by the Ministry of Education, Taiwan, R.O.C. under the ATU plan.
Author information
Authors and Affiliations
Corresponding author
Additional information
GenBank accession number for the 16S rRNA gene sequence of strain CC-Bw-6T is KC820654.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lin, SY., Hameed, A., Wen, CZ. et al. Lysobacter lycopersici sp. nov., isolated from tomato plant Solanum lycopersicum . Antonie van Leeuwenhoek 107, 1261–1270 (2015). https://doi.org/10.1007/s10482-015-0419-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-015-0419-1