Abstract
The litter-degrading dung fungus Coprinopsis cinerea has the high number of seventeen different laccase genes. In this work, ten different monokaryons were compared in their ability to produce laccases in two different complete media at different temperatures. Few strains showed laccase activity at the optimal growth temperature of 37 °C. Nine of the strains gave laccase activities between 0.2 and 5.9 U mL−1 at the suboptimal temperature of 25 °C in mKjalke medium. Laccase activities in YMG/T medium were detected for only three strains (0.5–4.5 U mL−1). Zymograms of supernatants from mKjalke medium resulted in total in 10 different laccase bands but strains differed in distribution. LC–MS/MS analysis with Mascot searches of the annotated C. cinerea genome identified isoenzymes from five different genes (Lcc1, Lcc2, Lcc5, Lcc9 and Lcc10) and of Lcc1 three and of Lcc5 two distinct electrophoretical forms. Lcc1 and Lcc5 were expressed in all laccase positive strains, but not all forms were found in all of the strains. Lcc2, Lcc9 and Lcc10 occurred only in three strains as minor laccases, indicating that Lcc1 and Lcc5 are the main laccases of C. cinerea secreted in liquid mKjalke medium.
Similar content being viewed by others
Introduction
Laccases (EC 1.10.3.2, benzenediol:oxygen oxidoreductase) are four copper-atoms containing oxidases capable of oxidising phenolic and other aromatic compounds (Leonowicz et al. 2001). In nature, these multi-copper oxidases occur widespread in the fungal kingdom, particularly in basidiomycetes (Hoegger et al. 2006). In many higher fungi, laccase genes exist in larger families that code for different isoenzymes with phenoloxidase activities (Kilaru et al. 2006a; Courty et al. 2009; Martinez et al. 2009; Floudas et al. 2012; Olson et al. 2012; Pezzella et al. 2012). Mainly laccases of straw- and wood-degrading species have been studied with respect to production, biochemical characteristics and potential biotechnological uses (Baldrian 2006). In contrast, biological functions of laccases usually remain speculative (Kües and Rühl 2011).
Paralogous laccase genes can be differentially regulated (Piscitelli et al. 2011). Laccase production in higher basidiomycetes is frequently under nutritional control (Dong et al. 2005; Stajić et al. 2006; Teerapatsakul et al. 2007), or it is stress-induced (Jaszek et al. 2006), developmentally regulated (Zhao and Kwan 1999) or part of a defence reaction (Hiscox et al. 2010), or it occurs in presence of specific inducers such as copper or potential phenolic substrates such as 2,4-dimethylphenol, ferulic acid, syringic acid and vanillin (Piscitelli et al. 2011) or during growth on lignocellulosic substrates (Stajić et al. 2006; Rühl et al. 2008). For routine fungal cultivation in laboratories, laccases are usually produced in simple liquid media (e.g. see Heinzkill et al. 1998; Ikehata et al. 2004; Han et al. 2005; Tong et al. 2007).
Often in liquid culture, more than one enzyme is produced. These may be isoenzymes coming from different genes or isoforms coming from a same gene and differing from each other due to differential splicing or by posttranslational modifications (Palmieri et al. 1997, 2000; Dong et al. 2005; D’Souza-Ticlo et al. 2006; Linke et al. 2005; Lorenzo et al. 2006; Giardina et al. 2007). In some wood-degrading Trametes species, 20 different isoenzymes and/or isoforms of specific laccases were detected (Dong et al. 2005) and strains of the edible white-rot mushroom Pleurotus ostreatus have been reported to produce isoenzymes from at least 4 distinct laccase genes (summarised by Pezzella et al. 2009).
The litter-degrading ink-cap mushroom Coprinopsis cinerea has in total 17 different laccase genes which present one of the largest groups of laccase genes ever described for a fungus (Kilaru et al. 2006a). Sixteen of the genes (lcc1 to lcc14, lcc16 and lcc17) translate into full-length proteins and 15 of these full-length proteins (all but Lcc8) have a typical secretion signal for release into the environment (Hoegger et al. 2004; Kilaru et al. 2006a). So far, hardly anything is known about expression of these multiple laccase genes. Schneider et al. (1999) isolated laccase CcL (synonymous to Lcc1) as the only enzyme from an unnamed C. cinerea culture broth. N-terminal sequencing allowed to assign CcL to a subcloned cDNA of gene lcc1 used for heterologous laccase production in Aspergillus oryzae. Evidence for expression of two other genes (lcc2, lcc3) is available from a cDNA library produced for subcloning laccase genes from mycelium of strain IFO 8371 grown at 26 °C in rich soy-bean-flour-based FG4 medium (Yaver et al. 1999).
Identification of different laccase isoenzymes and isoforms from culture supernatants is usually quite laborious. Classically, enzymes have individually to be purified and sequenced. For this, high amounts of laccase are required and, in consequence, such approaches are restricted to only a few species and for most species the number of expressed isoenzymes and -forms remains unknown. In contrast, with available fungal genomes new proteomic techniques help in definition of enzymes, even when present in minor amounts. In this study, we used LC–MS/MS with Mascot searches of the annotated C. cinerea genome published by Stajich et al. (2010) to identify laccases secreted in liquid culture by a collection of monokaryotic C. cinerea strains of different genetic backgrounds. Results show that media composition, growth temperature and the strain used influenced the outcome of laccase production. In modified Kjalke (mKjalke) medium at 25 °C under optimum yields of enzymatic activities, 9 of 10 analysed strains produced laccases Lcc1 and Lcc5 whereas some of the strains in addition gave also different forms of Lcc2, Lcc9 and/or Lcc10.
Materials and methods
Coprinopsis cinerea strains and culture conditions
All strains used in this study (see Table 1) were monokaryons of C. cinerea and well established laboratory strains (Kertesz-Chaloupková et al. 1998). Some of the strains are fully unrelated to each other (such as H5, JV6, Okayama 7 #130), others with common mating type genes and/or the trp-1.1,1.6 mutant gene share parts of their genomes. Yeast and malt extract used were from Oxoid (Basingstoke, UK), agar from Serva (Heidelberg, Germany). For stock cultures, fungi were grown at 37 °C on YMG/T medium (per litre: 4 g yeast extract, 10 g malt extract, 4 g glucose, 0.1 g tryptophan; solidified with 1 % agar when required) as a classical complete medium for cultivation of C. cinerea (YMG; Rao and Niederpruem 1969) to which tryptophan is added in order to allow also trp-auxotrophic strains to grow well (Granado et al. 1997). For precultures in liquid medium, ten mycelial agar plugs of Ø 6 mm were transferred into 500 mL flasks filled with either 50 mL YMG/T or 50 mL of mKjalke medium [i.e. per litre: 10 g yeast extract, 20 g glucose, 0.5 g CaCl2 × 2 H2O, 2 g KH2PO4, 50 mg MgSO4 × 7 H2O; adapted from Kjalke et al. (1992) as modified by Rühl and Kües (2009)]. Flasks were incubated for 4 days at 37 °C as standing cultures. Afterwards, precultures were homogenised by an Ultra-Turrax® (IKA Werke GmbH & Co. KG, Staufen, Germany) for about 30 s at 8,000 rpm followed by 30 s at 9,500 rpm. 5 mL of homogenised mycelium was used for inoculation of main-cultures in 500 mL flasks with 100 mL of YMG/T or mKjalke medium, each supplemented with 0.1 mM CuSO4. Strains were cultivated for 12 or more days on a rotary shaker at 120 rpm (agitation helps growth and laccase production in liquid medium) at suboptimal (25 °C) and optimal (37 °C) temperature for vegetative growth of the species (Kües 2000), respectively. Per experimental set-up and per culture condition, at least three parallel cultures were analysed. At least every second day, 1 mL aliquots were collected from the shaken cultures and stored at −20 °C for further analysis.
Laccase activity assay
In standard assay, laccase activity was determined at room temperature with 5 mM ABTS [2,2′-azino-bis-(3-ethylbenzthiazoline-6-sulfonate); AppliChem GmbH, Darmstadt, Germany] in 100 mM sodium acetate buffer at pH 5.0 at which laccases of C. cinerea tend to be most stable (unpublished results). Oxidation of ABTS into its cation radical (ABTS•+) was measured by an increase of absorbance at 420 nm (ε = 36,000 M−1 cm−1) for 5 min (Johannes and Majcherczyk 2000). One unit of activity was defined as the amount of enzyme needed to oxidise 1 μmol ABTS per min and activities are given in U per volume.
SDS-PAGE
Proteins were separated by SDS-PAGE in a Multigel-Long chamber of Biometra GmbH (Göttingen, Germany) using 4 % stacking and 12 % resolving gels (Laemmli 1970). In preliminary experiments, laccases were found to be not denatured by SDS, but the method allowed higher resolution of the zymograms compared to native PAGE (unpublished results).
Defrosted supernatant samples were concentrated with Vivaspin 2 (10.000 MWCO) ultrafiltration units (Sartorius GmbH, Göttingen, Germany). Aliquots of concentrated supernatants equal to 40 mU of laccase activity, diluted in deionised water to 12.5 μL and mixed 1:1 with loading buffer [0.06 M Tris–HCl (pH 6.8), 2 % SDS, 10 % glycerol, 0.025 % bromphenol blue (w/v)], were loaded per well for zymograms, whereas each 12.5 μL of the concentrated supernatants mixed 1:1 with loading buffer were used for Coomassie-stained gels. Electrophoresis was conducted at a constant current of 15 mA until the samples reached the resolving gel and continued at 25 mA for migration of proteins within the resolving gel.
For native laccase staining, gels were washed in 100 mM sodium acetate buffer (pH 5.0) and then incubated with 10 mM MBTH (3-methyl-2-benzothiazolinone-hydrazone hydrochloride) and 10 mM DHPPA (3,4-dihydroxyhydrocinnamic acid) dissolved in the same buffer, until protein bands showed up. For Coomassie staining, gels were fixed in 12 % (w/v) trichloroacetic acid (TCA) for at least 1 h and stained overnight in colloidal Coomassie solution [(10 % (v/v) phosphoric acid, 10 % (w/v) ammonium sulfate, 0.12 % (w/v) Coomassie Brilliant Blue G250 (Serva, Heidelberg, Germany) in water/methanol (80/20, v/v) solution]. Gels were washed with water until an optimal contrast between bands and background levels were obtained.
Protein identification
Protein bands of Coomassie stained gels were cut with a razor blade and proteins within gel pieces were digested with trypsin as described by Havliš et al. (2003) with modifications of Blödner et al. (2007). The tryptic peptides were dissolved in 2 % formic acid and separated on a 12 cm capillary column (ID 180 μm) packed with 3 μm particles of Reprosil-Pur C18-AQ (Dr. Maisch GmbH, Ammerbuch, Germany). Peptides were analysed by LC-ESI–MS/MS (LC 1100, Agilent; Esquire 3000, Bruker Daltonik GmbH, Bremen, Germany) and resulting raw data with DataAnalysis v. 3.0 software (Bruker Daltonik GmbH). Proteins were identified by searching the data against a database of the annotated genome of C. cinerea Okayama 7 #130 (Stajich et al. 2010) combined with the SwissProt database (http://us.expasy.org/sprot/), using a local installation of Mascot 2.2 software (www.matrixscience.com). The following settings were used for the database search: C-carbamidomethylation as fixed modification, M-oxidation as variable modification, peptide mass tolerance 1.4 Da, peptide charge 1+, 2+, and 3+, MS/MS tolerance 0.4 Da, and 1 missed cleavage allowed.
Results
Laccase activities depend on strain, medium and temperature
Previous tests of monokaryon FA2222 indicated that the strain at the optimal growth temperature of 37 °C produced no laccase in liquid YMG/T cultures (Kilaru et al. 2006b). YMG/T contains only 0.43 μM copper ions, too little to supply larger amounts of laccase molecules with each 4 copper atoms (Kilaru et al. 2006b). Therefore, in this study we added 0.1 mM CuSO4 to the medium, a concentration which did not negatively affect the growth of the fungus. However, copper supplementation had little effect on all tested strains in the production of active laccase in liquid YMG/T at 37 °C (Table 2). Most strains had no or only insignificant laccase activities. Noticeable activities of 0.13 and 0.09 U mL−1 were only measured for strains 306 and Okayama 7 #130, respectively.
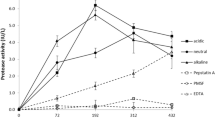
When reducing the growth temperature to suboptimal 25 °C, laccase activities for strain 306 did not alter but the two strains 218 and PG78 gave acceptable enzymatic activities in the culture supernatants (Table 2). In the experiment shown in Fig. 1, strain 218 and strain PG78 cultures showed first laccase activity at day 4, respectively day 5 of cultivation. The cultures had a maximum activity of 4.49 ± 0.16 U mL−1 at day 6, respectively of 2.35 ± 1.51 U mL−1 at day 7 of cultivation to then slowly decrease in activity to finally 3.55 ± 0.13 U mL−1, respectively 1.83 ± 1.53 U mL−1 at day 12 of cultivation. The overall laccase activity curves for the two strains at 25 °C were reproducible between different sets of experiments. Enzymatic activities in parallel cultures of strain PG78 were generally more variable than in cultures of strain 218 (see Fig. 1 for one set of experiments).
Laccase activity of C. cinerea strains PG78 and 218 grown over a period of 12 days at 25 °C in liquid YMG/T medium supplemented with 0.1 mM CuSO4. Average values and standard deviations have been calculated from measurements of each three parallel cultures. For better clarity of the presentation and avoiding confusion by overlapping lines, standard deviations are only shown in one direction
mKjalke medium with 10 g yeast-extract and 20 g glucose is richer in both N- and C-sources compared to YMG/T medium that contains 4 g of nitrogen-rich yeast extract (10.9 % nitrogen of total weight), 10 g nitrogen-pour malt extract (1.1 % nitrogen of total weight) and 10 g glucose. As in previous cultivation in YMG/T, 0.1 mM CuSO4 was added to the mKjalke medium and all C. cinerea monokaryons were cultivated at 37 °C and at 25 °C. At the optimal growth temperature at 37 °C, laccase production in the CuSO4-supplemented mKjalke medium for most strains was negligible or there were no detectable laccase activities in the culture supernatants (Table 2). In contrast, at 25 °C all but one strain (monokaryon LN118) gave acceptable amounts of laccase activities (Table 2). For most of the strains, laccase activity curves followed a similar pattern over the time (Fig. 2). First enzyme activities in mKjalke medium at 25 °C were encountered at day 4 of cultivation. Highest activities were obtained at later stages at days 6–10 of cultivation (Fig. 2), with best yields for monokaryons AT8 (5.88 ± 0.39 U mL−1), 218 (3.42 ± 0.68 U mL−1), 306 (2.91 ± 0.10 U mL−1), and FA2222 (2.81 ± 1.39 U mL−1). The activity curve of monokaryon FA2222 differed from others by late appearance of laccase activities, starting at day 8 of cultivation (Fig. 2b). Also in mKjalke, laccase activities between different sets of experiments were usually reproducible. Only strains PG78 and FA2222 showed higher variability in enzyme activities in parallel cultures (Fig. 2 and not shown).
Laccase activities of C. cinerea strains FA2222, H5, JV6 (a) and 218, 316, AT8 and PG78 (b) grown over a period of 12 days, respectively 13 days for FA2222, at 25 °C in mKjalke medium supplemented with 0.1 mM CuSO4. Average values and standard deviations have been calculated from measurements of each three parallel cultures. For better clarity of the presentation and avoiding confusion by overlapping lines, standard deviations are only shown in one direction per curve (either above or below)
Levels of laccase activities of strains 218 and PG78 at 25 °C in YMG/T and mKjalke medium were comparable (Table 2). MBTH and DHPPA co-stained zymograms of culture supernatants of strain 218 showed that the main bands of laccase activities were identical in the two media as well as over the time of cultivation within a medium (Fig. 3a). Similarly, in supernatants of monokaryon PG78, independently of culture medium and day of laccase activity, the main activity was always attributed to the same band at a lower position in the SDS-PAGE gels. Other bands at higher positions were of different intensity in the YMG/T and mKjalke samples or completely missing in supernatants of the other medium (Fig. 3b). In different series of cultivation of the strains, the same sets of bands were observed but with variations in intensities (not shown). The lower band was always most prominent for both strains and in both media.
Native PAGE of supernatants of C. cinerea strains 218 (a) and PG78 (b) grown at 25 °C in mKjalke and YMG/T medium. 40 mU of laccase were loaded of cultivation days 5, 7, 9 and 11. Subsequent to electrophoresis, the gel was stained with the laccase substrates MBTH and DHPPA until bands showed up. 20 mU (as measured by ABTS under our standard test conditions) of purified Lcc5 (Kilaru et al. unpublished results) was used as an internal standard. Note that the intensity of staining of the band of purified Lcc5 is similar to the corresponding major band in the fungal culture supernatants
Different isoenzymes account for the laccase activity of different strains
Supernatants from 25 °C cultures of C. cinerea strains grown in mKjalke with laccase activities of about 1 U mL−1 and higher as measured with ABTS (Table 2) were concentrated up to 30 fold and concentrated culture supernatants were loaded for native gel electrophoresis either for establishing a zymogram by detection of enzymatic activities (40 mU laccase per well) or for Coomassie staining of the complete secretome (12.5 μL concentrated culture supernatant per well, representing 450–1,300 mU of laccase).
The MBTH and DHPPA co-stained zymograms for all strains differed from each other in total numbers and positions of bands (between 4 and 7) and in intensities of bands shared between strains. In total by position, 10 different band types were defined (Fig. 4, Table 3). Only two bands (numbers 8 and 10) were present in all strains. Deduced from the staining intensity, in all strains either one or both of these bands were major bands of laccase activity with respect to co-staining by MBTH and DHPPA. Furthermore, bands 5, 6 and 7 were other main bands in monokaryons 306, AT8 and JV6, respectively (Fig. 4). Positions of main laccase activities in the zymograms corresponded with distinct bands in parallel Coomassie-stained gels, whereas bands were not evident at most positions of minor enzymatic activities (not further shown). For protein identification for all strains, corresponding bands or regions (i.e. where in Coomassie-stained gels no visible band existed) were cut from the Coomassie-stained gels and gel pieces were treated with trypsin. In case of strain 306, bands 6 and 7 had particular strong intensities in the activity-stained gels and were overlapping with each other (Fig. 4). Therefore, an extra band 6/7 was cut for this strain from the zone of Coomassie-stained gels in order to better differentiate specific laccases in band 6 and 7.
Native SDS-PAGE of concentrated supernatants of different C. cinerea strains grown for 7–9 days (see Table 2 for the harvest of each specific strain) at 25 °C in mKjalke medium. Per lane, samples of 40 mU of laccase activity were loaded. After electrophoresis, the gel was stained with MBTH and DHPPA. Numbers 1–10 define laccase bands of different positions
For the mostly faint higher bands at gel positions 1–4 (Fig. 4), significant peptide hits to laccases (ion score >43) were only obtained in case of the most intensive band 2 of strain H5 (see Supplementary Table S1). Laccases Lcc1 and Lcc5, respectively, were detected (Table 3). In the faintly stained band 2 of monokaryon 306, a single peptide of Lcc5 was found (see Supplementary Table S1).
Bands of higher intensity at gel positions 5–10 (Fig. 4) in nearly all instances gave significant results to specific laccases of Mascot protein score >100 (see Supplementary Table S1). In all strains, Lcc1 was detected as the only laccase in band 8. Lcc5 was detected as the only laccase in band 10 of all monokaryons but strain H5, where in addition Lcc1 was found. Furthermore, Lcc1 was present in band 9 of strain AT8. In conclusion, Lcc1 was present in two bands of strain AT8 and in three bands of strain H5, and Lcc5 was present in two bands of strains 306 and H5 (see Table 3). Minor laccases in strains 306, AT8 and JV6 were Lcc2, Lcc9 and/or Lcc10. Lcc2 was detected with a high Mascot protein score (284) in band 6 of strain 306 and one peptide for Lcc2 was detected in the corresponding laccase-active band 6 of strain JV6 (Table 3). Lcc9 was detected in band 6/7 of strain 306 and in band 5 of strain AT8. Lcc10 was present in band 6/7 and band 7 of strain 306 and in band 7 of strain AT8. Altogether, strains 306 and AT8 had the highest number of different laccases, i.e. 5 and 4 isoenzymes, respectively.
In most of the analysed samples, also several other secreted proteins were detected with significant Mascot protein scores (not further shown). Notably, no other enzymes than laccases were detected that could account for phenoloxidase activity in the zymograms. All peptides detected for any of the laccases (see Supplementary Table S1) are specific to the individual enzymes so that any cross-identification was excluded.
Discussion
In this study, we have shown that laccase secretion in monokaryotic C. cinerea strains depends on the cultivation medium and the cultivation temperature, with highest activities obtained in mKjalke medium at 25 °C. Of various possible culture media, C. cinerea grows best at 37 °C in YMG medium (Rao and Niederpruem 1969; Kües 2000). Under such optimal growth conditions (under extra addition of 0.5 mM tryptophan and 0.1 mM CuSO4), nearly all of our tested strains did not produce any laccase. Thus, the pure addition of copper to YMG does not induce laccase production, unlike in various typical white rot species (Ceriporiopsis subvermispora, Coriolopsis rigida, P. ostreatus, Lentinus tigrinus, Trametes spp.; Palmieri et al. 2000; Galhaup and Haltrich 2001; Saparrat et al. 2002; Manubens et al. 2007; Tong et al. 2007; Shutova et al. 2008; Lebrun et al. 2011). When decreasing the growth temperature to 25 °C, enzymatic activity significantly increased in cultures of the two C. cinerea strains 218 and PG78. The effect by changing the growth temperature from optimal 37 °C to suboptimal 25 °C was much more dramatic when using mKjalke medium for cultivation. All but one strain had high laccase activities at 25 °C (Table 2). Possibly, this lower suboptimal growth temperature stresses the fungus and laccase might be produced as a response (Kües and Rühl 2011). In many other basidiomycetes, optimum growth temperatures also do not correspond well with best enzyme production. For Trametes versicolor, the highest laccase activity was recorded by Šnajdr and Baldrian (2007) at a temperature of 35 °C although the optimum growth temperature of the fungus is 28 °C (Xavier et al. 2007). Similarly, P. ostreatus produces laccase best at 30 °C but grows best at 24 °C (Šnajdr and Baldrian 2007). The optimum temperatures for growth and laccase production of Cyathus bulleri are 24 °C and 30 °C, respectively (Vasdev et al. 2005). Interestingly, a Trametes sp. isolate produced highest amounts of laccase at 28 °C in a glucose medium and at 37 °C in cellobiose medium (Tong et al. 2007). Our experiments showed that the medium is also of importance for laccase yields in C. cinerea. mKjalke medium is richer in total resources than YMG/T but actual C/N ratios may play a role in regulation. Fungi differ in nutritional regulation. Low nitrogen has been reported in Pycnoporus cinnabarinus to induce laccase production (Eggert et al. 1996) whereas in P. ostreatus and Lentinula edodes higher nitrogen amounts caused better yields in laccase activity (Kaal et al. 1995). Apart from total amounts of nitrogen, the type of nitrogen source can be of influence on laccase production as shown in P. ostreatus and T. versicolor (Kaal et al. 1995; Mikiashvili et al. 2005, 2006).
Fungi may secrete different laccase isoenzymes under different nutritious and inductive conditions (Muñoz et al. 1997; Téllez-Téllez et al. 2005). For native production of laccases in basidiomycete fungi, the nutrient composition of the growth medium is very important, as this might have an impact on the secretion pattern of active enzymes (Muñoz et al. 1997; Pointing et al. 2000; Teerapatsakul et al. 2007). Dong et al. (2005) for example showed that the maximum activity and isoenzyme/isoform patterns in supernatants of Trametes gallica cultures depended on the used media as well as on the incubation condition (shaken or static). Altogether, the authors found 20 different laccase bands, three of which occurred in all types of cultures.
T. versicolor secreted laccases LacI and LacII in different isoenzyme ratios in liquid culture, depending on the lignocellulosic wastes used for cultivation (Moldes et al. 2004). Also two isoenzymes were present in cultures of Trametes sp. AH28-2 that were differently regulated by aromatic inducers (Xiao et al. 2004). Pleurotus pulmonarius has at least three laccase isoenzymes/isoforms, one of which was present in non-induced cultures, another one only in ferulic acid- and vanillin-induced cultures and the third in non-induced and induced cultures (De Souza et al. 2004). In P. ostreatus, three isoenzymes (two with their genes identified: i.e. POXA1b from gene lacc6 and POXC from gene lacc10) were found in culture supernatants, either produced constitutively or induced by supplemented Cu2+ or wheat straw extract (Palmieri et al. 2000; Castanera et al. 2012). Furthermore in copper supplemented cultures in this fungus, the heterodimeric laccase isoforms POXA3a and POXA3b exist due to differential splicing of transcripts of gene lacc2 and dimerisation with either one of two alternative small protein subunits of 16 and 18 kDa that distinguish from each other by one amino acid and by glycosylation and come from two genes not related to laccase genes (Giardina et al. 2007). Genes for other biochemically described laccases of P. ostreatus (POXA1w, POXA2) still await identification (Castanera et al. 2012). In this work, we detected by LC–MS/MS analysis the five different laccases Lcc1, Lcc2, Lcc5, Lcc9 and Lcc10 and Lcc1 in three and Lcc5 in two different forms (Table 3). The results of this study show that at least five of the in total 17 laccase genes of C. cinerea (Kilaru et al. 2006a) are functionally expressed. The occurrence of proteins Lcc1 and Lcc5 in bands of different positions within the gels (Fig. 4) suggests different posttranslational modifications or also homo- or heterodimerization to occur with the enzymes.
Little is known about the biological functions of laccases and why different isoenzymes and different isoforms are produced by many fungal species (Kües and Rühl 2011). Deduced from the presented data, Lcc1 and Lcc5 are universal laccases in cultures of C. cinerea, while other minor laccases (Lcc2, Lcc9 and Lcc10) might rather be strain-specific. The seventeen C. cinerea genes for laccases subdivide into two evolutionary distinct families (Kilaru et al. 2006a). All laccases detected in this study belong to laccase subfamily I. Of the detected laccases, Lcc5, Lcc9 and Lcc10 form a phylogenetic clade of closely related enzymes whereas Lcc1 and Lcc2 are diverged from this group and to each other (Kilaru et al. 2006a). Lcc5 shows 66/78 %, respectively 64/75 % identity/similarity to Lcc9 and Lcc10, but only 53/66 % identity/similarity to Lcc1. Lcc2 is similar distantly related to Lcc1 (53/66 % identity/similarity) and to Lcc5 (49/65 % identity/similarity). Most dissimilar between Lcc1 and Lcc5 are their substrate binding domains (Hoegger et al. 2004; Kilaru et al. 2006a). Currently it is not known, how much expression under shared physiological conditions reflects similarities in enzymatic properties and specific biological functions. Biochemical data of Lcc1 and data on Lcc5 suggest that the enzymes are not absolutely replaceable with each other (Schneider et al. 1999; Kilaru et al. unpublished results). Lcc1 is more effective than Lcc5 in oxidising classical laccase substrates but Lcc5 is more stable. Better stability may be a reason why Lcc5 was for all strains the most prominent band under all culturing conditions (Figs. 3 and 4).
References
Baldrian P (2006) Fungal laccases—occurrence and properties. FEMS Microbiol Rev 30:215–242
Blödner C, Majcherczyk A, Kües U, Polle A (2007) Early drought-induced changes to the needle proteome of Norway spruce. Tree Physiol 27:1423–1431
Castanera R, Pérez G, Omarini A, Alfaro M, Pisabarro AG, Faraco V, Amore A, Ramírez L (2012) Transcriptional and enzymatic profiling of Pleurotus ostreatus laccase genes in submerged and solid-state fermentation cultures. Appl Environ Microbiol 78:4037–4045
Courty PE, Hoegger PJ, Kilaru S, Kohler A, Buée M, Garbaye J, Martin F, Kües U (2009) Phylogenetic analysis, genomic organization, and expression analysis of multi-copper oxidases in the ectomycorrhizal basidiomycete Laccaria bicolor. New Phytol 182:736–750
D’Souza-Ticlo D, Verma AK, Mathew M, Raghukumar C (2006) Effect of nutrient nitrogen on laccase production, its isozyme pattern and effluent decolorization by the fungus NIOCC #2a, isolated from mangrove wood. Indian J Mar Sci 35:364–372
De Souza CGM, Tychanowicz GK, De Souza DF, Peralta RM (2004) Production of laccase isoforms by Pleurotus pulmonarius in response to presence of phenolic and aromatic compounds. J Basic Microbiol 44:129–136
Dong JL, Zhang YW, Zhang RH, Huang WZ, Zhang YZ (2005) Influence of culture conditions on laccase production and isozyme patterns in the white-rot fungus Trametes gallica. J Basic Microbiol 45:190–198
Eggert C, Temp U, Eriksson KEL (1996) The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: purification and characterization of the laccase. Appl Environ Microbiol 62:1151–1158
Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martínez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Górecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kües U, Kumar TKA, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, Rokas A, Ruiz-Dueñas FJ, Sabat G, Salamov A, Samejima M, Schmutz J, Slot JC, St. John F, Stenlid J, Sun H, Sun S, Syed K, Tsang A, Wiebenga A, Young D, Pisabarro A, Eastwood DC, Martin F, Cullen D, Grigoriev IV, Hibbett DS (2012) The paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719
Galhaup C, Haltrich D (2001) Enhanced formation of laccase activity by the white-rot fungus Trametes pubescens in the presence of copper. Appl Microbiol Biotechnol 56:225–232
Giardina P, Autore F, Faraco V, Festa G, Palmieri G, Piscitelli A, Sannia G (2007) Structural characterization of heterodimeric laccases from Pleurotus ostreatus. Appl Microbiol Biotechnol 75:1293–1300
Granado JD, Kertesz-Chaloupková K, Aebi M, Kües U (1997) Restriction enzyme-mediated DNA integration in Coprinus cinereus. Mol Gen Genet 256:28–36
Han GK, Park SH, Kim SH, Park HG, Park WM (2005) Detection and recovery of hydrolytic enzymes from spent compost of four mushroom species. Folia Microbiol 50:103–106
Havliš J, Thomas H, Šebela M, Shevchenko A (2003) Fast-response proteomics by accelerated in-gel digestion of proteins. Anal Chem 75:1300–1306
Heinzkill M, Bech L, Halkier T, Schneider P, Anke T (1998) Characterization of laccases and peroxidases from wood-rotting fungi (family Coprinaceae). Appl Environ Microbiol 64:1601–1606
Hiscox J, Baldrian P, Rogers HJ, Boddy L (2010) Changes in oxidative enzyme activity during interspecific mycelial interactions involving the white-rot fungus Trametes versicolor. Fungal Genet Biol 47:562–571
Hoegger PJ, Navarro-González M, Kilaru S, Hoffmann M, Westbrook ED, Kües U (2004) The laccase gene family in Coprinopsis cinerea (Coprinus cinereus). Curr Genet 45:9–18
Hoegger PJ, Kilaru S, James TY, Thacker JR, Kües U (2006) Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J 273:2308–2326
Ikehata K, Buchanan ID, Smith DW (2004) Extracellular peroxidase production by Coprinus species from urea-treated soil. Can J Microbiol 50:57–60
Jaszek M, Grzywnowicz K, Malarczyk E, Leonowicz A (2006) Enhanced extracellular laccase activity as a part of the response system of white rot fungi: Trametes versicolor and Abortiporus biennis to paraquat-caused oxidative stress conditions. Pest Biochem Physiol 85:147–154
Johannes C, Majcherczyk A (2000) Laccase activity tests and laccase inhibitors. J Biotechnol 78:193–199
Kaal EEJ, Field JA, Joyce TW (1995) Increasing ligninolytic enzyme activities in several white-rot basidiomycetes by nitrogen-sufficient media. Biores Technol 53:133–139
Kertesz-Chaloupková K, Walser PJ, Granado JD, Aebi M, Kües U (1998) Blue light overrides repression of asexual sporulation by mating type genes in the basidiomycete Coprinus cinereus. Fungal Genet Biol 23:95–109
Kilaru S, Hoegger PJ, Kües U (2006a) The laccase multi-gene family in Coprinopsis cinerea has seventeen different members that divide into two distinct subfamilies. Curr Genet 50:45–60
Kilaru S, Hoegger PJ, Majcherczyk A, Bruns C, Shishido K, Bailey A, Foster GD, Kües U (2006b) Expression of laccase gene lcc1 in Coprinopsis cinerea under control of various basidiomycete promoters. Appl Microbiol Biotechnol 71:200–210
Kjalke M, Andersen MB, Schneider P, Christensen B, Schülein M, Welinder KG (1992) Comparison of structure and activities of peroxidases from Coprinus cinereus, Coprinus macrorhizus and Arthromyces ramosus. Biochim Biophys Acta 1120:248–256
Kües U (2000) Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol Mol Biol Rev 64:316–353
Kües U, Rühl M (2011) Multiple multi-copper oxidase gene families in basidiomycetes—what for? Curr Genomics 12:72–94
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lebrun JD, Lamy I, Mougin C (2011) Favouring the bioavailability of Zn and Cu to enhance the production of lignin-modifying enzymes in Trametes versicolor cultures. Biores Technol 102:3103–3109
Leonowicz A, Cho NS, Luterek J, Wilkolazka A, Wojtaś-Wasilewska M, Matuszewska A, Hofrichter M, Wesenberg D, Rogalski J (2001) Fungal laccase: properties and activity on lignin. J Basic Microbiol 41:185–227
Linke D, Bouws H, Peters T, Nimtz M, Berger RG, Zorn H (2005) Laccases of Pleurotus sapidus: characterization and cloning. J Agric Food Chem 53:9498–9505
Lorenzo M, Moldes D, Sanromán MA (2006) Effect of heavy metals on the production of several laccase isoenzymes by Trametes versicolor and on their ability to decolourise dyes. Chemosphere 63:912–917
Manubens A, Canessa P, Folch C, Avila M, Salas L, Vicuna R (2007) Manganese affects the production of laccase in the basidiomycete Ceriporiopsis subvermispora. FEMS Microbiol Lett 275:139–145
Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek C, Ferreira P, Ruiz-Duenas F, Martinez A, Kersten P, Hammel K, Vanden Wymelenberg A, Gaskell J, Lindquist E, Sabat G, BonDurant SS, Larrondo L, Canessa P, Vicuna R, Yadav J, Doddapaneni H, Subramanian V, Pisabarro A, Lavín J, Oguiza J, Master E, Henrissat B, Coutinho P, Harris P, Magnuson J, Baker S, Bruno K, Kenealy W, Hoegger PJ, Kües U, Ramalya P, Lucas S, Salamov A, Shapiro H, Tu H, Chee C, Misra M, Xie G, Teter S, Yaver D, James T, Mokrejs M, Pospisek M, Grigoriev I, Brettin T, Rokhsar D, Berka R, Cullen D (2009) Genome, transcriptome, and secretome of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci USA 106:1954–1959
Mikiashvili N, Elisashvili V, Wasser S, Nevo E (2005) Carbon and nitrogen sources influence the ligninolytic enzyme activity of Trametes versicolor. Biotechnol Lett 27:955–959
Mikiashvili N, Wasser SP, Nevo E, Elisashvili V (2006) Effects of carbon and nitrogen sources on Pleurotus ostreatus ligninolytic enzyme activity. World J Microbiol Biotechnol 22:999–1002
Moldes D, Lorenzo M, Sanromán MA (2004) Different proportions of laccase isoenzymes produced by submerged cultures of Trametes versicolor grown on lignocellulosic wastes. Biotechnol Lett 26:327–330
Muñoz C, Guillén F, Martínez AT, Martínez MJ (1997) Laccase isoenzymes of Pleurotus eryngii: characterization, catalytic properties, and participation in activation of molecular oxygen and Mn2+ oxidation. Appl Environ Microbiol 63:2166–2174
Olson Å, Aerts A, Asiegbu F, Belbahri L, Bouzid O, Broberg A, Canbäck B, Coutinho PM, Cullen D, Dalman K, Deflorio G, van Diepen LT, Dunand C, Duplessis S, Durling M, Gonthier P, Grimwood J, Fossdal CG, Hansson D, Henrissat B, Hietala A, Himmelstrand K, Hoffmeister D, Högberg N, James TY, Karlsson M, Kohler A, Kües U, Lee Y, Lin Y, Lind M, Lindquist E, Lombard V, Lucas S, Lundén K, Morin E, Murat C, Park J, Raffaello T, Rouzé P, Salamov A, Schmutz J, Solheim H, Ståhlberg J, Vélëz H, de Vries RP, Wiebenga A, Woodward S, Yakovlev I, Garbelotto M, Martin F, Grigoriev IV, Stenlid J (2012) Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol 194:1001–1013
Palmieri G, Giardina P, Bianco C, Scaloni A, Capasso A, Sannia G (1997) A novel white laccase from Pleurotus ostreatus. J Biol Chem 272:31301–31307
Palmieri G, Giardina P, Bianco C, Fontanella B, Sannia G (2000) Copper induction of laccase isoenzymes in the ligninolytic fungus Pleurotus ostreatus. Appl Environ Microbiol 66:920–924
Pezzella C, Autore F, Giardina P, Piscitelli A, Sannia G, Faraco V (2009) The Pleurotus ostreatus laccase multi-gene family: isolation and heterologous expression of new family members. Curr Genet 55:45–57
Pezzella C, Lettera V, Piscitelli A, Giardina P, Sannia G (2012) Transcriptional analysis of Pleurotus ostreatus laccase genes. Appl Microbiol Biotechnol. doi:10.1007/s00253-012-3980-9
Piscitelli A, Giardina P, Lettera V, Pezzella C, Sannia G, Faraco V (2011) Induction and transcriptional regulation of laccases in fungi. Curr Genomics 12:104–112
Pointing SB, Jones EBG, Vrijmoed LLP (2000) Optimization of laccase production by Pycnoporus sanguineus in submerged liquid culture. Mycologia 92:139–144
Rao PS, Niederpruem DJ (1969) Carbohydrate metabolism during morphogenesis of Coprinus lagopus (sensu Buller). J Bacteriol 100:1222–1228
Rühl M, Kües U (2009) Automated image analysis to observe pellet morphology in liquid cultures of filamentous fungi such as the basidiomycete Coprinopsis cinerea. Curr Trends Biotechnol Pharm 3:241–253
Rühl M, Fischer C, Kües U (2008) Ligninolytic enzyme activities alternate with mushroom production during industrial cultivation of Pleurotus ostreatus on wheat-straw-based substrate. Curr Trends Biotechnol Pharm 2:478–492
Saparrat MCN, Guillen F, Arambarri AM, Martinez AT, Martinez MJ (2002) Induction, isolation, and characterization of two laccases from the white rot basidiomycete Coriolopsis rigida. Appl Env Microbiol 68:1534–1540
Schneider P, Caspersen MB, Mondorf K, Halkier T, Skov LK, Østergaard PR, Brown KM, Brown SH, Xu F (1999) Characterization of a Coprinus cinereus laccase. Enzym Microb Tech 25:502–508
Shutova VV, Revin VV, Myakushina YA (2008) The effect of copper ions on the production of laccase by the fungus Lentinus (Panus) tigrinus. Appl Biochem Microbiol 44:619–623
Šnajdr J, Baldrian P (2007) Temperature affects the production, activity and stability of ligninolytic enzymes in Pleurotus ostreatus and Trametes versicolor. Folia Microbiol 52:498–502
Stajić M, Persky L, Friesem D, Hadar Y, Wasser SP, Nevo E, Vukojević J (2006) Effect of different carbon and nitrogen sources on laccase and peroxidases production by selected Pleurotus species. Enzym Microb Tech 38:65–73
Stajich JE, Wilke SK, Ahrén D, Au CH, Birren BW, Borodovsky M, Burns C, Canbäck B, Casselton LA, Cheng CK, Deng J, Dietrich FS, Fargo DC, Farmann ML, Gathman AC, Goldberg J, Guigó R, Hoegger PJ, Hooker JB, Huggins A, James TY, Kamada T, Kilaru S, Kodira C, Kües U, Kupfer D, Kwang HS, Lomsadze A, Lin W, Lilly WW, Ma LJ, Mackey AJ, Manning G, Martin F, Muraguchi H, Natvig DO, Palmerini H, Ramesh MA, Rehmeyer CJ, Roe BA, Shenoy N, Stankea M, Ter-Hovhannisyan V, Tunlid A, Velagapudi R, Visione TJ, Zeng Q, Zolan ME, Pukkila PJ (2010) Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc Natl Acad Sci USA 107:11889–11894
Teerapatsakul C, Abe N, Bucke C, Kongkathip N, Jareonkitmongkol S, Chitradon L (2007) Novel laccases of Ganoderma sp. KU-Alk4, regulated by different glucose concentration in alkaline media. World J Microbiol Biotechnol 23:1559–1567
Téllez-Téllez M, Sánchez C, Loera O, Díaz-Godínez G (2005) Differential patterns of constitutive intracellular laccases of the vegetative phase of Pleurotus species. Biotechnol Lett 27:1391–1394
Tong PG, Hong YZ, Xiao YZ, Zhang M, Tu XM, Cui TJ (2007) High production of laccase by a new basidiomycete, Trametes sp. Biotechnol Lett 29:295–301
Vasdev K, Dhawan S, Kapoor RK, Kuhad RC (2005) Biochemical characterization and molecular evidence of a laccase from the bird’s nest fungus Cyathus bulleri. Fungal Genet Biol 42:684–693
Xavier AMRB, Tavares APM, Ferreira R, Amado F (2007) Trametes versicolor growth and laccase induction with by-products of pulp and paper industry. Elect J Biotechnol 10:444–451
Xiao YZ, Chen Q, Hang J, Shi YY, Xiao YZ, Wu J, Hong YZ, Wang YP (2004) Selective induction, purification and characterization of a laccase isozyme from the basidiomycete Trametes sp. AH28-2. Mycologia 96:26–35
Yaver DS, Overjero MD, Xu F, Nelson BA, Brown KM, Halkier T, Bernauer S, Brown SH, Kauppinen S (1999) Molecular characterization of laccase genes from the basidiomycete Coprinus cinereus and heterologous expression of the laccase Lcc1. Appl Environ Microbiol 65:4943–4948
Zhao J, Kwan HS (1999) Characterization, molecular cloning, and differential expression analysis of laccase genes from the edible mushroom Lentinula edodes. Appl Environ Microbiol 65:4908–4913
Acknowledgments
We thank Karin Lange and Mojtaba Zomorrodi for their excellent technical support. Proteomics work was supported within the framework of a Common Lower Saxony-Israel-Project (ZN 2043) by the Ministry of Science and Culture of Lower Saxony.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rühl, M., Majcherczyk, A. & Kües, U. Lcc1 and Lcc5 are the main laccases secreted in liquid cultures of Coprinopsis cinerea strains. Antonie van Leeuwenhoek 103, 1029–1039 (2013). https://doi.org/10.1007/s10482-013-9883-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-013-9883-7