Abstract
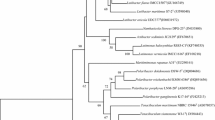
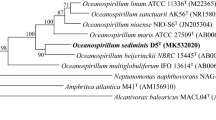
A novel Gram-negative, spiral shaped, motile bacterium, designated strain NIO-S6T, was isolated from a sediment sample collected from Off-shore Rameswaram, Tamilnadu, India. Strain NIO-S6T was found to be positive for oxidase, DNase and lysine decarboxylase activities and negative for catalase, gelatinase, lipase, ornithine decarboxylase, nitrate reductase, aesculinase, amylase and urease activities. The fatty acids were determined to be dominated by C10:0 3OH, C16:0, C16:1 and C18:1. Strain NIO-S6T contains Q-8 as the major respiratory quinone. The DNA G+C content of the strain NIO-S6T was determined to be 49.5 ± 0.6 mol %. Phylogenetic analysis based on 16S rRNA gene sequence of strain NIO-S6T indicated Oceanospirillum linum and Oceanospirillum maris of the family Oceanospirillaceae (phylum Proteobacteria) are the closest related species with sequence similarities of 98.4 and 97.8 % respectively. Other members of the family showed sequence similarities <96.4 %. However, DNA–DNA hybridization with Oceanospirillum linum LMG 5214T and Oceanospirillum maris LMG 5213T showed a relatedness of 31.5 and 46.9 % with respect to strain NIO-S6T. Based on the phenotypic characteristics and on phylogenetic inference, strain NIO-S6T is proposed as a novel species of the genus Oceanospirillum as Oceanospirillum nioense sp. nov. and the type strain is NIO-S6T (=MTCC 11154T = KCTC 32008T).

Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. Mol Biol 215:403–410
Brosius J, Palmer ML, Kennedy PJ, Noller HF (1978) Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA 75:4801–4805
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in Actinomycetes and Corynebacteria. J Gen Microbiol 100:221–230
Groth I, Schumann P, Rainey FA, Martin K, Schuetze B, Augsten K (1997) Demetria terragena gen. nov., sp. nov., a new genus of actinomycetes isolated from compost soil. Int J Syst Bacteriol 47:1129–1133
Guindon S, Lethiec F, Duroux P, Gascuel O (2005) PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33:W557–W559
Hylemon PB, Wells JS Jr, Krieg NR, Jannasch HW (1973) The genus Spirillum: a taxonomic study. Int J Syst Bacteriol 23:340–380
Johnson JL (1994) Similarity analysis of rRNAs. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 683–700
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Lányí B (1987) Classical and rapid identification methods for medically important bacteria. Methods Microbiol 19:1–67
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 3:208–218
Myers EW, Miller W (1988) Optimal alignments in linear space. Comput Appl Biosci 4:11–17
Pandey KK, Mayilraj S, Chakrabarti T (2002) Pseudomonas indica sp. nov., a novel butane-utilizing species. Int J Syst Evol Microbiol 52:1559–1567
Pot B, Gillis M (2005) Genus I. Oceanospirillum Hylemon, Wells, Krieg and Jannasch 1973, 361AL. In: Brenner DJ, Krieg NR, Staley JT (eds) Bergey’s manual of systematic bacteriology, 2nd edn, vol. 2. The Proteobacteria Part B The Gammaproteobacteria. Springer, New York, pp 271–281
Pot B, Gillis M, Hoste B, Van De Velde A, Bekaert F, Kersters K, De Ley J (1989) Intra- and intergeneric relationships of the genus Oceanospirillum. Int J Syst Bacteriol 39:23–34
Reddy GSN, Aggarwal RK, Matsumoto GI, Shivaji S (2000) Arthrobacter flavus sp. nov., a psychrophilic bacterium isolated from a pond in McMurdo Dry Valley Antarctica. Int J Syst Evol Microbiol 50:1553–1561
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Satomi M, Kimura B, Hamada T, Harayama S, Fujii T (2002) Phylogenetic study of the genus Oceanospirillum based on 16S rRNA and gyrB genes: emended description of the genus Oceanospirillum, description of Pseudospirillum gen. nov., Oceanobacter gen. nov. and Terasakiella gen. nov. and transfer of Oceanospirillum jannaschii and Pseudomonas stanieri to Marinobacterium as Marinobacterium jannaschii comb. nov. and Marinobacterium stanieri comb. nov. Int J Syst Evol Microbiol 52:739–747
Shivaji S, Ray MK, Saisree L, Jagannadham MV, Seshu Kumar G, Reddy GSN, Bhargava PM (1992) Sphingobacterium antarcticus sp. nov.: a psychrotrophic bacterium from the soils of Schirmacher Oasis, Antarctica. Int J Syst Bacteriol 42:102–116
Sly LI, Blackall LL, Kraat PC, Tian-Shen T, Sangkhobol V (1986) The use of second derivative plots for the determination of mol% guanine plus cytosine of DNA by the thermal denaturation method. J Microbiol Methods 5:139–156
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Terasaki Y (1979) Tranfer of five species and two subspecies of Spirillum to other genera (Aquaspirillum and Oceanospirillum), with emended descriptions of the species and subspecies. Int J Syst Bacteriol 29:130–144
Thompson JD, Higgins DG, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tourova TP, Antonov AS (1987) Identification of microorganisms by rapid DNA–DNA hybridisation. Methods Microbiol 19:333–355
Vandamme P, Pot B, Gillis M, De Vos P, Kersters K, Swings J (1996) Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev 60:407–438
Versalovic J, Koeuth T, Lupski JR (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucl Acids Res 19:6823–6831
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Truper HG (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Williams MA, Rittenberg SC (1957) A taxonomic study of the genus Spirillum Ehrenberg. Int Bull Bacteriol Nomen Tax. 7:49–112
Acknowledgments
We thank Dr. J. Euzeby for his expert suggestion for the correct species epithet and Latin etymology. We also thank Council of Scientific and Industrial Research (CSIR) and Department of Biotechnology, Government of India for financial assistance. We would like to thank Mr. Deepak for his help in 16S rRNA gene sequencing. The laboratory facility was extended by MMRF of NIO, RC, Kochi funded by the Ministry of Earth Sciences, New Delhi. TNRS is thankful to CSIR projects (SIP1302, SIP1308, OLP1201) other projects (GAP2131, SSP2490) for providing sample, facilities and funding. NIO contribution number is 5302.
Author information
Authors and Affiliations
Corresponding author
Additional information
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain NIO-S6T is JN205304.
All authors contributed equally to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Krishna, K.K., Bhumika, V., Thomas, M. et al. Oceanospirillum nioense sp. nov., a marine bacterium isolated from sediment sample of Palk bay, India. Antonie van Leeuwenhoek 103, 1015–1021 (2013). https://doi.org/10.1007/s10482-013-9881-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-013-9881-9