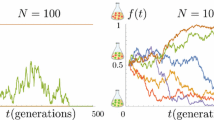
Abstract
Recurrent mutations are a common phenomenon in population genetics. They may be at the origin of the fixation of a new genotype, if they give a phenotypic advantage to the carriers of the new mutation. In this paper, we are interested in the genetic signature left by a selective sweep induced by recurrent mutations at a given locus from an allele \(A\) to an allele \(a\), depending on the mutation frequency. We distinguish three possible scales for the mutation probability per reproductive event, which entail distinct genetic signatures. Besides, we study the hydrodynamic limit of the \(A\)- and \(a\)-population size dynamics when mutations are frequent, and find non trivial equilibria leading to several possible patterns of polymorphism.







Similar content being viewed by others
References
Athreya, K.B., Ney, P.E.: Branching Processes. Dover, Mineola (2004). Reprint of the 1972 original (Springer, New York, MR0373040)
Baigent, S.: Lotka-Volterra Dynamics—an introduction. Preprint, University of College, London (2010)
Billiard, S., Smadi, C.: The interplay of two mutations in a population of varying size: a stochastic eco-evolutionary model for clonal interference. Stoch. Process. Appl. (2016). doi:10.1016/j.spa.2016.06.024
Billiard, S., Smadi, C.: Beyond clonal interference: scrutinizing the complexity of the dynamics of three competing clones. arXiv preprint arXiv:1607.04656 (2016)
Billiard, S., Collet, P., Ferrière, R., Méléard, S., Tran, V.C.: The effect of horizontal trait inheritance on competitive exclusion and coexistence. arXiv preprint arXiv:1511.09062
Brink-Spalink, R.: Stochastic Models in Population Genetics: The Impact of Selection and Recombination (2015)
Brink-Spalink, R., Smadi, C.: Genealogies of two linked neutral loci after a selective sweep in a large population of stochastically varying size. Adv. Appl. Probab. 49.1 (2017)
Cardano, G.: Ars Magna or the Rules of Algebra. Dover, New York (1968)
Champagnat, N.: A microscopic interpretation for adaptive dynamics trait substitution sequence models. Stoch. Process. Appl. 116(8), 1127–1160 (2006)
Champagnat, N., Méléard, S.: Polymorphic evolution sequence and evolutionary branching. Probab. Theory Relat. Fields 151(1–2), 45–94 (2011)
Champagnat, N., Jabin, P.-E., Méléard, S.: Adaptation in a stochastic multi-resources chemostat model. J. Math. Pures Appl. 101(6), 755–788 (2014)
Chan, Y.F., Marks, M.E., Jones, F.C., Villarreal, G., Shapiro, M.D., Brady, S.D., Southwick, A.M., Absher, D.M., Grimwood, J., Schmutz, J.: Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327(5963), 302–305 (2010)
Coville, J., Fabre, F.: Convergence to the equilibrium in a Lotka-Volterra ode competition system with mutations. arXiv preprint arXiv:1301.6237 (2013)
Cresko, W.A., Amores, A., Wilson, C., Murphy, J., Currey, M., Phillips, P., Bell, M.A., Kimmel, C.B., Postlethwait, J.H.: Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc. Natl. Acad. Sci. USA 101(16), 6050–6055 (2004)
Cuevas, J., Geller, R., Garijo, R., López-Aldeguer, J., Sanjuán, R.: Extremely high mutation rate of HIV-1 in vivo. PLoS Biol. 13(9), e1002251 (2015)
Diekmann, O.: A beginner’s guide to adaptive dynamics. Banach Cent. Publ. 63, 47–86 (2004)
Drake, J.W.: Rates of spontaneous mutation among RNA viruses. Proc. Natl. Acad. Sci. USA 90(9), 4171–4175 (1993)
Duffy, S., Shackelton, L.A., Holmes, E.C.: Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9(4), 267–276 (2008)
Dumortier, F., Llibre, J., Artés, J.C.: Qualitative Theory of Planar Differential Systems. Springer, Berlin (2006)
Erkko, H., Xia, B., Nikkilä, J., Schleutker, J., Syrjäkoski, K., Mannermaa, A., Kallioniemi, A., Pylkäs, K., Karppinen, S.-M., Rapakko, K.: A recurrent mutation in PALB2 in Finnish cancer families. Nature 446(7133), 316–319 (2007)
Ethier, S., Kurtz, T.: Markov Processes: Characterization and Convergence (1986)
Fournier, N., Méléard, S.: A microscopic probabilistic description of a locally regulated population and macroscopic approximations. Ann. Appl. Probab. 14(4), 1880–1919 (2004)
Gago, S., Elena, S.F., Flores, R., Sanjuán, R.: Extremely high mutation rate of a hammerhead viroid. Science 323(5919), 1308 (2009)
Griffiths, R.C.: Lines of descent in the diffusion approximation of neutral Wright-Fisher models. Theor. Popul. Biol. 17(1), 37–50 (1980)
Haldane, J.: The part played by recurrent mutation in evolution. Am. Nat. 67, 5–19 (1933)
Hermisson, J., Pennings, P.S.: Soft sweeps molecular population genetics of adaptation from standing genetic variation. Genetics 169(4), 2335–2352 (2005)
Hermisson, J., Pfaffelhuber, P.: The pattern of genetic hitchhiking under recurrent mutation. Electron. J. Probab. 13(68), 2069–2106 (2008)
Ikeda, N., Watanabe, S.: Stochastic Differential Equations and Diffusion Processes (1989)
Kimura, M., Ohta, T.: Stepwise mutation model and distribution of allelic frequencies in a finite population. Proc. Natl. Acad. Sci. USA 75(6), 2868–2872 (1978)
Messer, P.W., Petrov, D.A.: Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol. Evol. 28(11), 659–669 (2013)
Metz, J.A., Geritz, S.A., Meszéna, G., Jacobs, F.J., Van Heerwaarden, J.: Adaptive dynamics, a geometrical study of the consequences of nearly faithful reproduction. In: Stochastic and Spatial Structures of Dynamical Systems, vol. 45, pp. 183–231 (1996)
Moya, A., Holmes, E.C., González-Candelas, F.: The population genetics and evolutionary epidemiology of RNA viruses. Nat. Rev. Microbiol. 2(4), 279–288 (2004)
Nagylaki, T.: Evolution of a finite population under gene conversion. Proc. Natl. Acad. Sci. USA 80(20), 6278–6281 (1983)
Nair, S., Nash, D., Sudimack, D., Jaidee, A., Barends, M., Uhlemann, A.-C., Krishna, S., Nosten, F., Anderson, T.: Recurrent gene amplification and soft selective sweeps during evolution of multidrug resistance in malaria parasites. Mol. Biol. Evol. 24(2), 562–573 (2007)
Nohturfft, A., Hua, X., Brown, M.S., Goldstein, J.L.: Recurrent G-to-A substitution in a single codon of SREBP cleavage-activating protein causes sterol resistance in three mutant Chinese hamster ovary cell lines. Proc. Natl. Acad. Sci. USA 93(24), 13709–13714 (1996)
Pennings, P.S., Hermisson, J.: Soft sweeps iii: the signature of positive selection from recurrent mutation. PLoS Genet. 2(12), e186 (2006)
Pennings, P.S., Hermisson, J.: Soft sweeps ii: molecular population genetics of adaptation from recurrent mutation or migration. Mol. Biol. Evol. 23(5), 1076–1084 (2006)
Pokalyuk, C.: The effect of recurrent mutation on the linkage disequilibrium under a selective sweep. J. Math. Biol. 64(1–2), 291–317 (2012)
Prezeworski, M., Coop, G., Wall, J.D.: The signature of positive selection on standing genetic variation. Evolution 59(11), 2312–2323 (2005)
Repping, S., Skaletsky, H., Brown, L., van Daalen, S.K., Korver, C.M., Pyntikova, T., Kuroda-Kawaguchi, T., de Vries, J.W., Oates, R.D., Silber, S.: Polymorphism for a 1.6-mb deletion of the human y chromosome persists through balance between recurrent mutation and haploid selection. Nat. Genet. 35(3), 247–251 (2003)
Richard, M.: Limit theorems for supercritical age-dependent branching processes with neutral immigration. Adv. Appl. Probab. 43, 276–300 (2011)
Risheg, H., Graham, J.M., Clark, R.D., Rogers, R.C., Opitz, J.M., Moeschler, J.B., Peiffer, A.P., May, M., Joseph, S.M., Jones, J.R.: A recurrent mutation in MED12 leading to R961W causes Opitz-Kaveggia syndrome. Nat. Genet. 39(4), 451–453 (2007)
Scheinfeldt, L.B., Biswas, S., Madeoy, J., Connelly, C.F., Schadt, E.E., Akey, J.M.: Population genomic analysis of AL M S1 in humans reveals a surprisingly complex evolutionary history. Mol. Biol. Evol. 26(6), 1357–1367 (2009)
Schweinsberg, J., Durrett, R.: Random partitions approximating the coalescence of lineages during a selective sweep. Ann. Appl. Probab. 15(3), 1591–1651 (2005)
Shore, E.M., Xu, M., Feldman, G.J., Fenstermacher, D.A., Cho, T.-J., Choi, I.H., Connor, J.M., Delai, P., Glaser, D.L., LeMerrer, M.: A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat. Genet. 38(5), 525–527 (2006)
Skuce, P., Stenhouse, L., Jackson, F., Hypša, V., Gilleard, J.: Benzimidazole resistance allele haplotype diversity in United Kingdom isolates of teladorsagia circumcincta supports a hypothesis of multiple origins of resistance by recurrent mutation. Int. J. Parasitol. 40(11), 1247–1255 (2010)
Smadi, C.: An eco-evolutionary approach of adaptation and recombination in a large population of varying size. Stoch. Process. Appl. 125(5), 2054–2095 (2015)
Tishkoff, S.A., Reed, F.A., Ranciaro, A., Voight, B.F., Babbitt, C.C., Silverman, J.S., Powell, K., Mortensen, H.M., Hirbo, J.B., Osman, M.: Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 39(1), 31–40 (2007)
van Ditmarsch, D., Boyle, K.E., Sakhtah, H., Oyler, J.E., Nadell, C.D., Déziel, E., Dietrich, L.E.P., Xavier, J.B.: Convergent evolution of hyperswarming leads to impaired biofilm formation in pathogenic bacteria. Cell Rep. 4(4), 697–708 (2013)
Wilson, B.A., Petrov, D.A., Messer, P.W.: Soft selective sweeps in complex demographic scenarios. Genetics 198(2), 669–684 (2014)
Acknowledgements
The author would like to thank Helmut Pitters for fruitful discussions at the beginning of this work. She also wants to thank Jean-René Chazottes and Pierre Collet for advice and references on planar dynamical systems, as well as Pierre Recho for his help with the use of Mathematica, and two anonymous reviewers for their careful reading of the paper, and several suggestions and improvements. This work was partially funded by the Chair “Modélisation Mathémathique et Biodiversité” of Veolia Environnement—Ecole Polytechnique—Museum National d’Histoire Naturelle—Fondation X and the French national research agency ANR-11-BSV7-013-03.
Author information
Authors and Affiliations
Corresponding author
Appendix: Technical Results
Appendix: Technical Results
In this last section, we recall some results on birth and death processes whose proofs can be found in Lemma 3.1 in [44] and in [1], pp. 109 and 112.
Proposition A.1
Let \(Z=(Z_{t})_{t \geq 0}\) be a birth and death process with individual birth and death rates \(b\) and \(d \). For \(i \in \mathbb{Z}_{+}\), \(T_{i}=\inf \{ t\geq 0, Z_{t}=i \}\) and \(\mathbb{P}_{i}\) (resp. \(\mathbb{E}_{i}\)) is the law (resp. expectation) of \(Z\) when \(Z_{0}=i\). Then
-
For \((i,j,k) \in \mathbb{Z}_{+}^{3}\) such that \(j \in (i,k)\),
$$ \mathbb{P}_{j}(T_{k}< T_{i})= \frac{1-(d/b)^{j-i}}{1-(d/b)^{k-i}}. $$(A.1) -
If \(d\neq b \in \mathbb{R}_{+}^{*}\), for every \(i\in \mathbb{Z}_{+}\) and \(t \geq 0\),
$$ \mathbb{P}_{i}(T_{0}\leq t )= \biggl( \frac{d(1-e^{(d-b)t})}{b-de^{(d-b)t}} \biggr)^{i}. $$(A.2) -
If \(0< d< b\), on the non-extinction event of \(Z\), which has a probability \(1-(d/b)^{Z_{0}}\), the following convergence holds:
$$ T_{N}/\log N \mathop{\to }_{N \to \infty } (b-d)^{-1}, \quad \textit{a.s.} $$(A.3)
The next lemma quantifies the time spent by a birth and death process with logistic competition in a vicinity of its equilibrium size. It is stated in [9], Theorem 3(c).
Lemma A.1
Let \(b,d,c\) be in \(\mathbb{R}_{+}^{*}\) such that \(b-d>0\). Denote by \((W_{t},t \geq 0)\) a density dependent birth and death process with birth rate \(bn\) and death rate \((d+c n/K)n\), where \(n \in \mathbb{N}\) is the current state of the process and \(K \in \mathbb{N}\) is the carrying capacity. Fix \(0 < \eta_{1} < (b - d)/c\) and \(\eta_{2} > 0\), and introduce the stopping time
Then, there exists \(V > 0\) such that, for any compact subset \(C\) of \(](b - d)/c - \eta_{1}, (b - d)/ c + \eta_{2} [\),
We end this section with a coupling of birth and death processes with close birth and death rates.
Lemma A.2
Let \(Z_{1}\) and \(Z_{2}\) be two birth and death processes with initial state 1 and respective individual birth and death rates \((b_{1},d _{1})\) and \((b_{2},d_{2})\) belonging to a common compact set \(D\) in \(\mathbb{R}_{+}^{2}\). Then we can couple \(Z_{1}\) and \(Z_{2}\) in such a way that
where the positive constant \(c\) only depends on \(D\).
Proof
For the sake of simplicity, we assume in the proof than \(b_{1}< b_{2}\) and \(d_{1}< d_{2}\), but other cases can be treated similarly. Let \(B(ds,d\theta )\) and \(D(ds,d\theta )\) be two independent Poisson random measures with intensity \(ds d\theta \). We can construct the two processes \(Z_{1}\) and \(Z_{2}\) with respect to the measures \(B\) and \(D\). For \(i \in \{1,2\}\), introduce
We also introduce an auxiliary birth and death process, \(Z_{3}\), with individual birth and death rates \((b_{3},d_{3})=(b_{2},d_{1})\), which will allow us to compare \(Z_{1}\) and \(Z_{2}\):
As \(b_{1}< b_{2}\) and \(d_{1}< d_{2}\), we have the following almost sure inequalities:
Applying Itô’s Formula yields that \(M_{i}(t)= Z_{i}(t)e^{-(b_{i}-d _{i})t}\), \(i \in \{1,2,3\}\) are martingales, and we can express the quadratic variation of their differences. Using (A.6) we get:
Then, by taking the expectation, we obtain
where for the first inequality we have used that the square of a martingale is a submartingale, and in the last one the continuity of the functions involved. We obtain similarly
But applying (A.6) we get that
and that
which yields
Taking the expectation and reasoning similarly as before give for every positive \(t\),
Using that for \(a,b,c\geq 0\), \((a-c)^{2} \leq 2(a-b)^{2}+2(b-c)^{2} \) and adding (A.7) end the proof of Lemma A.2. □
Rights and permissions
About this article
Cite this article
Smadi, C. The Effect of Recurrent Mutations on Genetic Diversity in a Large Population of Varying Size. Acta Appl Math 149, 11–51 (2017). https://doi.org/10.1007/s10440-016-0086-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10440-016-0086-x
Keywords
- Eco-evolution
- Birth and death process with immigration
- Selective sweep
- Coupling
- Competitive Lotka-Volterra system with mutations