Abstract
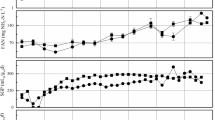
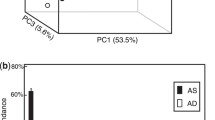
To identify potential linkages between specific bacterial populations and process performance in anaerobic digestion, the dynamics of bacterial community structure was monitored with high-throughput sequencing in triplicate anaerobic digesters treating animal waste. Firmicutes and Bacteroidetes were found as the two most abundant populations, however, with contrasting population dynamics in response to organic overloading. Firmicutes dominated the bacterial community during stable process performance at low organic loading rate, representing over 50 % of the bacterial abundance. In contrast, the onset of organic overloading raised the relative abundance of Bacteroidetes from 20 ± 2.6 to 44 ± 3.1 %. In addition to the significant negative correlation between the relative abundance of Firmicutes and Bacteroidetes, populations of Firmicutes and Bacteroidetes were found to be linked to process parameters including organic loading rate, volatile fatty acids concentration, and methane production. Therefore, the population abundance ratio of Firmicutes to Bacteroidetes (F/B ratio) was suggested as a potential indicator for process performance. The interactions between Firmicutes and Bacteroidetes populations could be exploited to develop strategies for the prevention of performance perturbation in anaerobic digestion processes.






Similar content being viewed by others
References
Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N (2011) Examining the global distribution of dominant archaeal populations in soil. ISME J 5:908–917
Briones A, Coats E, Brinkman C (2014) Should we build “obese” or “lean” anaerobic digesters? PLoS One 9:97252
Briones A, Raskin L (2003) Diversity and dynamics of microbial communities in engineered environments and their implications for process stability. Curr Opin Biotechnol 14:270–276
Callaway TR, Dowd SE, Edrington TS, Anderson RC, Krueger N, Bauer N, Kononoff PJ, Nisbet DJ (2010) Evaluation of bacterial diversity in the rumen and feces of cattle fed different levels of dried distillers grains plus solubles using bacterial tag-encoded FLX amplicon pyrosequencing. J Anim Sci 88:3977–3983
Chao A (1984) Nonparametric estimation of the number of classes in a population. Scand J Stat 11:265–270
Chao A, Shen T-J (2003) Nonparametric estimation of Shannon’s index of diversity when there are unseen species in sample. Environ Ecol Stat 10:429–443
Chen S, He Q (2015) Persistence of Methanosaeta populations in anaerobic digestion during process instability. J Ind Microbiol Biotechnol 42:1129–1137
Chen S, Zamudio Cañas EM, Zhang Y, Zhu Z, He Q (2012) Impact of substrate overloading on archaeal populations in anaerobic digestion of animal waste. J Appl Microbiol 113:1371–1379
Chen Y, Cheng JJ, Creamer KS (2008) Inhibition of anaerobic digestion process: a review. Bioresour Technol 99:4044–4064
Colwell RK, Coddington JA (1994) Estimating terrestrial biodiversity through extrapolation. Philos Trans R Soc Lond Ser B Biol Sci 345:101–118
Fulthorpe RR, Roesch LFW, Riva A, Triplett EW (2008) Distantly sampled soils carry few species in common. ISME J 2:901–910
Girvan MS, Campbell CD, Killham K, Prosser JI, Glover LA (2005) Bacterial diversity promotes community stability and functional resilience after perturbation. Environ Microbiol 7:301–313
Grabowski A, Tindall BJ, Bardin V, Blanchet D, Jeanthon C (2005) Petrimonas sulfuriphila gen. nov., sp nov., a mesophilic fermentative bacterium isolated from a biodegraded oil reservoir. Int J Syst Evol Microbiol 55:1113–1121
Hashsham SA, Fernandez AS, Dollhopf SL, Dazzo FB, Hickey RF, Tiedje JM, Criddle CS (2000) Parallel processing of substrate correlates with greater functional stability in methanogenic bioreactor communities perturbed by glucose. Appl Environ Microbiol 66:4050–4057
Ito T, Yoshiguchi K, Ariesyady HD, Okabe S (2011) Identification of a novel acetate-utilizing bacterium belonging to Synergistes group 4 in anaerobic digester sludge. ISME J 5:1844–1856
Kampmann K, Ratering S, Kramer I, Schmidt M, Zerr W, Schnell S (2012) Unexpected stability of Bacteroidetes and Firmicutes communities in laboratory biogas reactors fed with different defined substrates. Appl Environ Microbiol 78:2106–2119
Krakat N, Schmidt S, Scherer P (2011) Potential impact of process parameters upon the bacterial diversity in the mesophilic anaerobic digestion of beet silage. Bioresour Technol 102:5692–5701
Liu FH, Wang SB, Zhang JS, Zhang J, Yan X, Zhou HK, Zhao GP, Zhou ZH (2009) The structure of the bacterial and archaeal community in a biogas digester as revealed by denaturing gradient gel electrophoresis and 16S rDNA sequencing analysis. J Appl Microbiol 106:952–966
Mata-Alvarez J, Macé S, Llabrés P (2000) Anaerobic digestion of organic solid wastes. An overview of research achievements and perspectives. Bioresour Technol 74:3–16
McGarvey JA, Miller WG, Zhang R, Ma Y, Mitloehner F (2007) Bacterial population dynamics in dairy waste during aerobic and anaerobic treatment and subsequent storage. Appl Environ Microbiol 73:193–202
Mnif S, Zayen A, Karray F, Bru-Adan V, Loukil S, Godon JJ, Chamkha M, Sayadi S (2012) Microbial population changes in anaerobic membrane bioreactor treating landfill leachate monitored by single-strand conformation polymorphism analysis of 16S rDNA gene fragments. Int Biodeterior Biodegrad 73:50–59
Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S (2012) Host-gut microbiota metabolic interactions. Science 336:1262–1267
Pavlostathis SG, Giraldo-Gomez E (1991) Kinetics of anaerobic treatment: a critical review. Crit Rev Environ Control 21:411–490
Peet RK (1974) The measurement of species diversity. Annu Rev Ecol Syst 5:285–307
Riviere D, Desvignes V, Pelletier E, Chaussonnerie S, Guermazi S, Weissenbach J, Li T, Camacho P, Sghir A (2009) Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J 3:700–714
Schink B (1997) Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev 61:262–280
Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310
Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Svensson BH, Sørensen SJ, Karlsson A (2013) 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol 85:612–626
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Zhang XJ, Yue SQ, Zhong HH, Hua WY, Chen RJ, Cao YF, Zhao LP (2011) A diverse bacterial community in an anoxic quinoline-degrading bioreactor determined by using pyrosequencing and clone library analysis. Appl Microbiol Biotechnol 91:425–434
Zhang Y, He Q (2013) Characterization of bacterial diversity in drinking water by pyrosequencing. Water Sci Technol Water Supply 13:358–367
Zhang Y, Zhang XL, Zhang HW, He Q, Zhou QX, Su ZC, Zhang CG (2009) Responses of soil bacteria to long-term and short-term cadmium stress as revealed by microbial community analysis. Bull Environ Contam Toxicol 82:367–372
Zhu Z, Hsueh MK, He Q (2011) Enhancing biomethanation of municipal waste sludge with grease trap waste as a co-substrate. Renew Energy 36:1802–1807
Ziganshin A, Schmidt T, Scholwin F, Il’inskaya O, Harms H, Kleinsteuber S (2011) Bacteria and archaea involved in anaerobic digestion of distillers grains with solubles. Appl Microbiol Biotechnol 89:2039–2052
Acknowledgments
This work was partly supported by a US Environmental Protection Agency Grant XA-83539201 and the Science Alliance—Tennessee Center of Excellence. SC was partly supported by the Institute for a Secure and Sustainable Environment at the University of Tennessee, Knoxville.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, S., Cheng, H., Wyckoff, K.N. et al. Linkages of Firmicutes and Bacteroidetes populations to methanogenic process performance. J Ind Microbiol Biotechnol 43, 771–781 (2016). https://doi.org/10.1007/s10295-016-1760-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-016-1760-8