Abstract
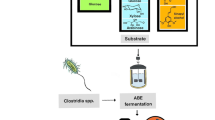
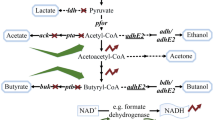
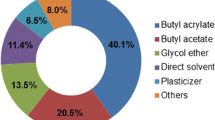
With the incessant fluctuations in oil prices and increasing stress from environmental pollution, renewed attention is being paid to the microbial production of biofuels from renewable sources. As a gasoline substitute, butanol has advantages over traditional fuel ethanol in terms of energy density and hygroscopicity. A variety of cheap substrates have been successfully applied in the production of biobutanol, highlighting the commercial potential of biobutanol development. In this review, in order to better understand the process of acetone–butanol–ethanol production, traditional clostridia fermentation is discussed. Sporulation is probably induced by solvent formation, and the molecular mechanism leading to the initiation of sporulation and solventogenesis is also investigated. Different strategies are employed in the metabolic engineering of clostridia that aim to enhancing solvent production, improve selectivity for butanol production, and increase the tolerance of clostridia to solvents. However, it will be hard to make breakthroughs in the metabolic engineering of clostridia for butanol production without gaining a deeper understanding of the genetic background of clostridia and developing more efficient genetic tools for clostridia. Therefore, increasing attention has been paid to the metabolic engineering of E. coli for butanol production. The importation and expression of a non-clostridial butanol-producing pathway in E. coli is probably the most promising strategy for butanol biosynthesis. Due to the lower butanol titers in the fermentation broth, simultaneous fermentation and product removal techniques have been developed to reduce the cost of butanol recovery. Gas stripping is the best technique for butanol recovery found so far.



Similar content being viewed by others
References
Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, Chou KJ, Hanai T, Liao JC (2007) Metabolic engineering of Escherichia coli for 1-butanol production. Metab Eng 10:305–311
Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched- chain higher alcohols as biofuels. Nature 451:86–89
Bennett GN, Rudolph FB (1995) The central metabolic pathway from acetyl-CoA to butyryl-CoA in Clostridium acetobutylicum. FEMS Microbiol Rev 17:241–249
Borden JR, Papoutsakis ET (2007) Dynamics of genomic-library enrichment and identification of solvent tolerance genes for Clostridium acetobutylicum. Appl Environ Microbiol 73:3061–3068
Boynton ZL, Bennett GN, Rudolph FB (1996) Cloning, sequencing, and expression of clustered genes encoding β-hydroxybutyryl-coenzyme A (CoA) dehydrogenase, crotonase, and butyryl-CoA dehydrogenase from Clostridium acetobutylicum ATCC 824. J Bacteriol 178:3015–3024
Bryant DL, Blaschek HP (1988) Buffering as a means for increasing growth and butanol production by Clostridium acetobutylicum. J Ind Microbiol 3:49–55
Chen CK, Blaschek HP (1999) Acetate enhances solvent production and prevents degeneration in Clostridium beijerinckii BA101. Appl Microbiol Biotechnol 52:170–173
Dabrock B, Bahl H, Gottschalk G (1992) Parameters affecting solvent production by Clostridium pasteurianum. Appl Environ Microbiol 58:1233–1239
Desai RP, Papoutsakis ET (1999) Antisense RNA strategies for metabolic engineering of Clostridium acetobutylicum. Appl Environ Microbiol 65:936–945
Dürre P (2007) Biobutanol: an attractive biofuel. Biotechnol J 2:1525–1534
Dürre P, Böhringer M, Nakottel S, Schaffer S, Thormann K, Zickner B (2002) Transcriptional regulation of solventogenesis in Clostridium acetobutylicum. J Mol Microbiol Biotechnol 4:295–300
Dürre P, Hollergschwandner C (2004) Initiation of endospore formation in Clostridium acetobutylicum. Anaerobe 10:69–74
Evans PJ, Wang HY (1988) Enhancement of butanol formation by Clostridium acetobutylicum in the presence of decanol-oleyl alcohol mixed extractants. Appl Environ Microbiol 54:1662–1667
Ezeji TC, Karcher PM, Qureshi N, Blaschek HP (2005) Improving performance of a gas stripping-based recovery system to remove butanol from Clostridium beijerinckii fermentation. Bioprocess Biosyst Eng 27:207–214
Ezeji TC, Qureshi N, Blaschek HP (2003) Production of acetone, butanol and ethanol by Clostridium beijerinckii BA101 and in situ recovery by gas stripping. World J Microbiol Biotechnol 19:595–603
Ezeji TC, Qureshi N, Blaschek HP (2004) Acetone butanol ethanol (ABE) production from concentrated substrate: reduction in substrate inhibition by fed-batch technique and product inhibition by gas stripping. Appl Microbiol Biotechnol 63:653–658
Ezeji TC, Qureshi N, Blaschek HP (2005) Continuous butanol fermentation and feed starch retrogradation: butanol fermentation sustainability using Clostridium beijerinckii BA101. J Biotechnol 115:179–187
Ezeji TC, Qureshi N, Blaschek HP (2007) Bioproduction of butanol from biomass: from genes to bioreactors. Curr Opin Biotechnol 18:220–227
Ezeji TC, Qureshi N, Blaschek HP (2007) Production of acetone butanol (AB) from liquefied corn starch, a commercial substrate, using Clostridium beijerinckii coupled with product recovery by gas stripping. J Ind Microbiol Biotechnol 34:771–777
Feustel L, Nakotte S, Dürre P (2004) Characterization and development of two reporter gene systems for Clostridium acetobutylicum. Appl Environ Microbiol 70:798–803
Garcia A, Lannotti EL, Fischer JL (1986) Butanol fermentation liquor production and separation by reverse osmosis. Biotechnol Bioeng 28:785–791
Girbal L, Mortier-Barriére I, Raynaud F, Rouanet C, Croux C, Soucaille P (2003) Development of a sensitive gene expression reporter system and an inducible promoter–repressor system for Clostridium acetobutylicum. Appl Environ Microbiol 69:4985–4988
Glieder A, Farinas ET, Arnold FH (2002) Laboratory evolution of a soluble, self-sufficient, highly active alkane hydroxylase. Nat Biotechnol 20:1135–1139
Green EM, Bennett GN (1996) Inactivation of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC824. Appl Biochem Biotechnol 57–58:213–221
Green EM, Boynton ZL, Harris LM, Rudolph FB, Papoutsakis ET, Bennett GN (1996) Genetic manipulation of acid formation pathways by gene inactivation in Clostridium acetobutylicum ATCC 824. Microbiology 142:2079–2086
Harris LM, Blank L, Desai RP, Welker NE, Papoutsakis ET (2001) Fermentation characterization and flux analysis of recombinant strains of Clostridium acetobutylicum with an inactivated solR gene. J Ind Microbiol Biotechnol 27:322–328
Harris LM, Welker NE, Papoutsakis ET (2002) Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC824. J Bacteriol 184:3586–3597
Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP (2007) The ClosTron: a universal gene knock-out system for the genus Clostridium. J Microbiol Methods 70:452–464
Hermann M, Fayolle F, Marchal R, Podvinl L, Sebald M, Vandecasteelei JP (1985) Isolation and characterization of butanol-resistant mutants of Clostridium acetobutylicum. Appl Environ Microbiol 50:1238–1243
Inui M, Suda M, Kimura S, Yasuda K, Suzuki H, Toda H, Yamamoto S, Okino S, Suzuki N, Yukawa H (2008) Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Appl Microbiol Biotechnol 77:1305–1316
Jones DT, Shirley M, Wu X, Keis S (2000) Bacteriophage infections in the industrial acetone butanol (AB) fermentation process. J Mol Microbiol Biotechnol 2:21–26
Kashket ER, Cao ZY (1993) Isolation of a degeneration-resistant mutant of Clostridium acetobutylicum NCIMB 8052. Appl Environ Microbiol 59:4198–4202
Lee J, Mitchell WJ, Tangney M, Blaschek HP (2005) Evidence for the presence of an alternative glucose transport system in Clostridium beijerinckii NCIMB 8052 and the solvent- hyperproducing mutant BA101. Appl Environ Microbiol 71:3384–3387
Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, Jung KS (2008) Fermentative butanol production by clostridia. Biotechnol Bioeng 101:209–228
Lienhardt J, Schripsema J, Qureshi N, Blaschek HP (2002) Butanol production by Clostridium beijericknii BA101 in an immobilized cell biofilm reactor. Appl Biochem Biotechnol 98–100:591–598
Liyanage H, Holcroft P, Evans VJ, Keis S, Wilkinson SR, Kashket ER, Young M (2000) A new insertion sequence, ISCb1, from Clostridium beijerinckii NCIMB 8052. J Mol Microbiol Biotechnol 2:107–113
Louis P, McCrae SL, Charrier C, Flint HJ (2007) Organization of butyrate synthetic genes in human colonic bacteria: phylogenetic conservation and horizontal gene transfer. FEMS Microbiol Lett 269:240–247
Maddox IS, Steiner E, Hirsch S, Wessner S, Gutierrez NA, Gapes JR, Schuster KC (2000) The cause of “acid crash” and “acidogenic fermentations” during the batch acetone-butanol-ethanol (ABE-) fermentation process. J Mol Microbiol Biotechnol 2:95–100
Mavrovouniotis ML (1990) Group contributions for estimating standard Gibbs energies of formation of biochemical compounds in aqueous solution. Biotechnol Bioeng 36:1070–1082
Mavrovouniotis ML (1991) Estimation of standard Gibbs energy changes of biotransformations. J Biol Chem 266:14440–14445
Mermelstein LD, Papoutsakis ET, Petersen DJ, Bennett GN (1993) Metabolic engineering of Clostridium acetobutylicum ATCC 824 for increased solvent production by enhancement of acetone formation enzyme-activities using a synthetic acetone operon. Biotechnol Bioeng 42:1053–1060
Mermelstein LD, Welker NE, Bennett GN, Papoutsakis ET (1992) Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824. Biotechnology 10:190–195
Milestone NB, Bibby DM (1981) Concentration of alcohols by adsorption on silicalite. J Chem Technol Biotechnol 31:732–736
Mutschlechner O, Swoboda H, Gapes JR (2000) Continuous two-stage ABE-fermentation using Clostridium beijerinckii NRRL B592 operating with a growth rate in the first stage vessel close to its maximal value. J Mol Microbiol Biotechnol 2:101–105
Nair RV, Green EM, Watson DE, Bennett GN, Papoutskis ET (1999) Regulation of the sol locus genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 by a putative transcriptional repressor. J Bacteriol 181:319–330
Nair RV, Papoutsakis ET (1994) Expression of plasmid-encoded aad in Clostridium acetobutylicum M5 restores vigorous butanol production. J Bacteriol 176:5843–5846
Nakayama S, Kosaka T, Hirakawa H, Matsuura K, Yoshino S, Furukawa K (2008) Metabolic engineering for solvent productivity by downregulation of the hydrogenase gene cluster hupCBA in Clostridium saccharoperbutylacetonicum strain N1–4. Appl Microbiol Biotechnol 78:483–493
Nakayama S, Morita T, Negishi H, Ikegami T, Sakaki K, Kitamoto D (2008) Candida krusei produces ethanol without production of succinic acid; a potential advantage for ethanol recovery by pervaporation membrane separation. FEMS Yeast Res 8(5):706–714
Narberhaus F, Giebeler K, Bahl H (1992) Molecular characterization of the dnaK gene region of Clostridium acetobutylicum, including grpE, dnaJ, and a new heat shock gene. J Bacteriol 174:3290–3299
Nölling J, Breton G, Omelchenko MV, Makarova KS, Zeng QD, Gibson R, Lee HM, Dubois J, Qiu D, Hitti J, Wolf YI, Tatusov RL, Sabathe F, Doucette-Stamm L, Soucaille P, Daly MJ, Bennett GN, Koonin EV, Smith DR (2001) Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J Bacteriol 183:4823–4838
Oh MK, Liao JC (2000) DNA microarray detection of metabolic responses to protein overproduction in Escherichia coli. Metab Eng 2:201–209
Parekh M, Blaschek HP (1999) Butanol production by hypersolvent-producing mutant Clostridium beijerinckii BA101 in corn steep water medium containing maltodextrin. Biotechnol Lett 21:45–48
Quixiey KWM, Reid SJ (2000) Construction of a reporter gene vector for Clostridium beijerinckii using a Clostridium endoglucanase gene. J Mol Microbiol Biotechnol 2:53–57
Qureshi N, Blaschek HP (2001) Recovery of butanol from fermentation broth by gas stripping. Renew Energy 22:557–564
Qureshi N, Saha BC, Hector RE, Hughes SR, Cotta MA (2008) Butanol production from wheat straw by simultaneous saccharification and fermentation using Clostridium beijerinckii: Part I—batch fermentation. Biomass Bioenerg 32:168–175
Rao G, Mutharasan R (1987) Altered electron flow in continuous cultures of Clostridium acetobutylicum induced by viologen dyes. Appl Environ Microbiol 53:1232–1235
Ravagnani A, Jennert KC, Steiner E, Grunberg R, Jefferies JR, Wilkinson SR, Young DI, Tidswell EC, Brown DP, Youngman P, Morris JG, Young M (2000) Spo0A directly controls the switch from acid to solvent production in solvent-forming Clostridia. Mol Microbiol 37:1172–1185
Santangelo JD, Kuhn A, Treuner-Lange A, Dürre P (1998) Sporulation and time course expression of sigma-factor homologous genes in Clostridium acetobutylicum. FEMS Microbiol Lett 161:157–164
Sauer U, Treuner A, Buchholz M, Santangelo JD, Dürre P (1994) Sporulation and primary sigma factor homologous genes in Clostridium acetobutylicum. J Bacteriol 176:6572–6582
Scotcher MC, Rudolph FB, Bennett GN (2005) Expression of abrB310 and sinR, and effects of decreased abrB310 expression on the transition from acidogenesis to solventogenesis, in Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 71:1987–1995
Shen CR, Liao JC (2008) Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via keto-acid pathways. Metab Eng 10:312–320
Shi ZP, Zhang CY, Chen JX, Mao ZG (2005) Performance evaluation of acetone–butanol continuous flash extractive fermentation process. Bioprocess Biosyst Eng 27:175–183
Shinto H, Tashiro Y, Kobayashi G, Sekiguchi T, Hanai T, Kuriya Y, Okamoto M, Sonomoto K (2008) Kinetic study of substrate dependency for higher butanol production in acetone-butanol-ethanol fermentation. Process Biochem 43:1452–1461
Sillers R, Chow A, Tracy AB, Papoutsakis ET (2008) Metabolic engineering of the non-sporulating, non-solventogenic Clostridium acetobutylicum strain M5 to produce butanol without acetone demonstrate the robustness of the acid-formation pathways and the importance of the electron balance. Metab Eng 10:321–332
Tashiro Y, Shinto H, Hayashi M, Baba S, Kobayashi G, Sonomoto K (2007) Novel high-efficient butanol production from butyrate by non-growing Clostridium saccharoperbutylacetonicum N1–4 (ATCC 13564) with methyl viologen. J Biosci Bioeng 104:238–240
Thormann K, Dürre P (2001) Orf5/SolR: a transcriptional repressor of the sol operon of Clostridium acetobutylicum? J Ind Microbiol Biotechnol 27:307–313
Thormann K, Feustel L, Lorenz K, Nakotte S, Dürre P (2002) Control of butanol formation in Clostridium acetobutylicum by transcriptional activation. J Bacteriol 184:1966–1973
Tomas CA, Welker NE, Papoutsakis ET (2003) Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell’s transcriptional program. Appl Environ Microbiol 69:4951–4965
Tummala SB, Junne SG, Papoutsakis ET (2003) Antisense RNA downregulation of coenzyme A transferase combined with alcohol-aldehyde dehydrogenase overexpression leads to predominantly alcohologenic Clostridium acetobutylicum fermentations. J Bacteriol 185:3644–3653
Tummala SB, Welker NE, Papoutsakis ET (2003) Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum. J Bacteriol 185:1923–1934
Vasconcelosi I, Girbal L, Soucaille P (1994) Regulation of carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH on mixtures of glucose and glycerol. J Bacteriol 176:1443–1450
Wiesenborn DP, Rudolph FB, Papoutsakis ET (1988) Thiolase from Clostridium acetobutylicum ATCC 824 and its role in the synthesis of acids and solvents. Appl Environ Microbiol 54:2717–2722
Wong J, Bennett GN (1996) The effect of novobiocin on solvent production by Clostridium acetobutylicum. J Ind Microbiol 16:354–359
Yan RT, Zhu CX, Golemboski C, Chen JS (1988) Expression of solvent-forming enzymes and onset of solvent production in batch cultures of Clostridium beijerinckii. Appl Environ Microbiol 54:642–648
Acknowledgments
We would like to acknowledge the financial support of the CAS 100 Talents Program (No. KGCXZ-YW-801).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zheng, YN., Li, LZ., Xian, M. et al. Problems with the microbial production of butanol. J Ind Microbiol Biotechnol 36, 1127–1138 (2009). https://doi.org/10.1007/s10295-009-0609-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-009-0609-9