Abstract
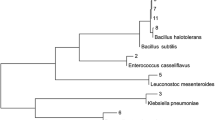
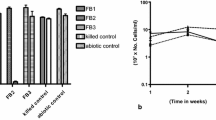
Shipping operations produce oily wastes that must be managed properly to avoid environmental pollution. The aim of this study was to characterize microorganisms occurring in ship bilge wastes placed in open lagoons and, particularly, to assess their potential to degrade polycyclic aromatic hydrocarbons (PAHs). A first-order kinetic was suitable for describing hydrocarbon biodegradation after 17 days of treatment. The calculated rate constants were 0.0668 and 0.0513 day−1 with a corresponding half-life of 10.3 and 13.5 days for the aliphatic and aromatic hydrocarbon fractions, respectively. At day 17, PAH removal percentages were: acenaphtylene 100, fluorene 95.2, phenanthrene 93.6, anthracene 70.3, and pyrene 71.5. Methyl phenanthrene removals were lower than that of their parent compound (3-methyl phenanthrene 83.6, 2-methyl phenanthrene 80.8, 1-methyl phenanthrene 77.3, 9-methyl phenanthrene 75.1, and 2,7-dimethyl phenanthrene 76.6). Neither pure cultures nor the microbial community from these wastes showed extracellular biosurfactant production suggesting that the addition of an exogenously produced biosurfactant may be important in enhancing hydrocarbon bioavailability and biodegradation. DNA analysis of bilge waste samples revealed a ubiquitous distribution of the nahAc genotype in the dump pools. Although almost all of the isolates grew on naphthalene as sole carbon source, only some of them yielded nahAc amplification under the experimental conditions used. The variety of PAHs in bilge wastes could support bacteria with multiple degradation pathways and a diversity of catabolic genes divergent from the classical nah-like type.






Similar content being viewed by others
References
Bosh R, Garcia-Valdes E, Moore ERB (1999) Genetic characterization and evolutionary implications of a chromosomally encoded naphthalene-degradation upper pathway from Pseudomonas stutzeri AN10. Gene 236:149–157
Commendatore MG, Esteves JL, Colombo JC (2000) Hydrocarbons in coastal sediments of Patagonia, Argentina: levels and probable sources. Mar Pollut Bull 40:989–998
Cooper DG (1986) Biosurfactants. Microbiol Sci 3145–3150
Cooper DG, Goldenberg BG (1987) Surface active agents from two Bacillus species. Appl Environ Microbiol 53:224–229
Ferrero M, Llobet-Brossa E, Lalucat J, García-Valdés E, Rosselló-Mora R, Bosch R (2002) Coexistence of two distinct copies of naphthalene degradation genes in Pseudomonas strains isolated from the western Mediterranean region. Appl Environ Microbiol 68:957–962
Gamati S, Gosselin C, Bergeron E, Chenier M, Truong TV, Bisaillon JG (1999) New plug flow slurry bioreactor for polycyclic aromatic hydrocarbon degradation. In: Alleman B, Leeson A (eds) Bioremediation technologies for polycyclic aromatic hydrocarbon compounds. Battelle Press, Columbus, Ohio, pp 1–6
Haigler BE, Gibson DT (1990) Purification and properties of NADH-ferredoxin NAP reductase, a component of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J Bacteriol 172:457–464
Irwin RJ, VanMouwerik M, Stevens L, Seese MD, Basham W (1997) Environmental contaminants encyclopedia. National Park Service, Water Resources Division, Fort Collins, Co.
Jeffrey AM, Yeh HJ, Jerina DM, Patel TR, Davey JF, Gibson DT (1975) Initial reactions in the oxidation of naphthalene by Pseudomonas putida. Biochemistry 14:575–584
Kanaly RA, Harayama S (2000) Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J Bacteriol 182:2059–2067
Karin A, Berry T, Burton DL (1997) Natural attenuation of diesel fuel in heavy clay soil. Can J Soil Sci 77:469–477
Lagrega MD, Buckingham PL, Evans JC (1994) Hazardous waste management. McGraw-Hill, New York
Laurie AD, Lloyd-Jones G (2000) Quantification of phnAc and nahAc in contaminated New Zealand soils by competitive PCR. Appl Environ Microbiol 66:1814–1817
Mills AL, Breuil C, Colwell RR (1978) Enumeration of petroleum-degrading marine and estuarine microorganisms by the most probable number method. Can J Microbiol 24:552–557
Moeseneder MM, Arrieta JM, Muyzer G, Winter C, Herndl GJ (1999) Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl Environ Microbiol 65:3518–3525
Morán AC, Olivera N, Commendatore M, Esteves JL, Siñeriz F (2000) Enhancement of hydrocarbon waste biodegradation by the addition of a biosurfactant from Bacillus subtilis O9. Biodegradation 11:65–71
Olivera N (2001) Biorremediación de sistemas contaminados con hidrocarburos. Universidad Nacional del Sur. Bahía Blanca, Argentina. PhD thesis
Olivera N, Commendatore MG, Morán AC, Esteves JL (2000) Biosurfactant-enhanced degradation of residual hydrocarbons from ship bilge wastes. J Ind Microbiol Biotechnol 25:70–73
Rosenberg E, Ron EZ (1999) High- and low-molecular-mass microbial surfactants. Appl Microbiol Biotechnol 52:154–162
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
Sepic E, Trier C, Leskovsek H (1996) Biodegradation studies of selected hydrocarbons from diesel oil. Analyst 121:1451–1456
Sepic E, Bricelj M, Leskovsek H (1997) Biodegradation studies of polyaromatic hydrocarbons in aqueous media. J Appl Microbiol 83:561–568
Stapleton RD, Sayler GS (1998) Assessment of the microbiological potential for the natural attenuation of petroleum hydrocarbons in a shallow aquifer system. Microb Ecol 36:349–361
Stapleton RD, Bright NG, Sayler GS (2000) Catabolic and genetic diversity of degradative bacteria from fuel-hydrocarbon contaminated aquifers. Microb Ecol 39:211–221
United Nations Environmental Programme (1992) Determinations of petroleum hydrocarbons in sediments. Reference methods for marine pollution studies, no. 20. UNEP, Nairobi, Kenya
Acknowledgements
This work was supported by the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and the Agencia Nacional de Promoción Científica y Tecnológica (PICT 07-04069. BID 1201/OC-AR), (PICT-PIP 4271/96), Argentina.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Olivera, N.L., Commendatore, M.G., Delgado, O. et al. Microbial characterization and hydrocarbon biodegradation potential of natural bilge waste microflora. J IND MICROBIOL BIOTECHNOL 30, 542–548 (2003). https://doi.org/10.1007/s10295-003-0078-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-003-0078-5