Abstract
A widening array of novel imaging biomarkers is being developed using ever more powerful clinical and preclinical imaging modalities. These biomarkers have demonstrated effectiveness in quantifying biological processes as they occur in vivo and in the early prediction of therapeutic outcomes. However, quantitative imaging biomarker data and knowledge are not standardized, representing a critical barrier to accumulating medical knowledge based on quantitative imaging data. We use an ontology to represent, integrate, and harmonize heterogeneous knowledge across the domain of imaging biomarkers. This advances the goal of developing applications to (1) improve precision and recall of storage and retrieval of quantitative imaging-related data using standardized terminology; (2) streamline the discovery and development of novel imaging biomarkers by normalizing knowledge across heterogeneous resources; (3) effectively annotate imaging experiments thus aiding comprehension, re-use, and reproducibility; and (4) provide validation frameworks through rigorous specification as a basis for testable hypotheses and compliance tests. We have developed the Quantitative Imaging Biomarker Ontology (QIBO), which currently consists of 488 terms spanning the following upper classes: experimental subject, biological intervention, imaging agent, imaging instrument, image post-processing algorithm, biological target, indicated biology, and biomarker application. We have demonstrated that QIBO can be used to annotate imaging experiments with standardized terms in the ontology and to generate hypotheses for novel imaging biomarker–disease associations. Our results established the utility of QIBO in enabling integrated analysis of quantitative imaging data.
Similar content being viewed by others
Introduction
Imaging, by CT and MR, has been ranked by physicians as the single most important medical innovation [1]. In addition to common clinical imaging modalities, biomedical research studies use many other rich and diverse types of imaging data, including high-resolution microscopy images, fluorescence imaging, and recently developed nanoparticle imaging. The biological significance of these imaging studies often involves identifying imaging biomarkers that are indicators of the underlying biology. Currently, quantitative imaging suffers from the lack of a standardized representation of quantitative image features and content [2–5].
As one resource that depends on this, the concept of “image biobanking” as an analog to tissue biobanking has great promise [6, 7]. Tools have become available for handling the complexity of genotype [8–12], and similar advancements are needed to describe phenotype, especially as derived from imaging [4, 13–20]. Publicly accessible resources that support large image archives support file sharing and have so far not yet merged into a framework that supports the collaborative work needed to meet the potential of quantitative image analysis. With the availability of tools for automatic ontology-based annotation of datasets with terms from biomedical ontologies, coupled with image archives and the means for batch selection and processing of image and clinical data, imaging will go through a similar increase in capability analogous to what advanced sequencing techniques have gone through in molecular biology.
Quantitative imaging biomarkers, which are the focus of this work, provide a numerical characterization of the underlying biology or pathology as opposed to using textual or categorical descriptions of an observer’s subjective visual interpretation. Accelerated by advances in imaging techniques, a wide array of novel quantitative imaging biomarkers have been developed and have demonstrated effectiveness in quantifying biological processes and in clinical use. For example, imaging biomarkers, such as change in tumor uptake measured by the standardized uptake value (SUV) of [18F]-FDG positron emission tomography (PET), can be used in monitoring disease progression, prediction of response to treatment, as well as drug development [21].
Fully describing a quantitative imaging biomarker and how it is used involves specifying a series of heterogeneous concepts that span the fields of imaging physics, contrast agent or probe chemistry, biology, and quantitation techniques. Interpretation of data used in the development and validation of quantitative imaging biomarkers requires these disparate concepts to be related together with scientifically rigorous epistemology. We posit that this motivates an immediate need for an ontology to represent the complex and heterogeneous imaging biomarker data and knowledge with sufficient coverage of these fields [22]. In this endeavor, we developed an ontology that represents the knowledge domain of quantitative imaging biomarkers and we have explored applications enabled by it.
Imaging biomarker research
The Biomarkers Definitions Working Group at National Institutes of Health defines biomarkers as characteristics that are objectively measured as indicators of normal biological processes, pathological changes, or pharmacologic responses to a therapeutic intervention [2, 23]. For example, the measurement of the serum concentration of the glycoprotein CA125 assayed by ELISA can be used to monitor therapy during treatment for ovarian cancer. When biomarkers are obtained from biomedical images, they are referred to as imaging biomarkers. While the term imaging biomarker is sometimes used to refer directly to the exogenous imaging agent or its molecular target, we use the term here in a broader context to represent the measurement of the characteristic obtained through imaging. There is much research interest in imaging biomarkers, and several related efforts have been initiated in imaging biomarker research in the scientific community.
The Radiological Society of North America has formed the Quantitative Imaging Biomarker Alliance (QIBA) to advance quantitative imaging in clinical care and facilitating imaging as a biomarker in clinical trials [24, 25]. While engaging many stakeholders across academia, government, and industry, the effort is limited to the most mature biomarkers. Also, they are not addressing biomarkers from a knowledge engineering perspective and have only limited activity associated with formal verification activities.
The Center for Biomarkers in Imaging at the Massachusetts General Hospital has developed a large imaging biomarker catalog with about 350 imaging biomarkers [26]. Although extensive, this cataloging effort is not focused on a structured and standardized representation of quantitative imaging biomarkers.
Biomedical Imaging Research
Imaging techniques allow non-invasive interrogation of the biology. This has fundamental utility in characterizing tissues and biological processes in the context of a living organism. The non-invasive nature also allows studies to be carried out serially, and thus enables characterization of biological changes over time.
Recently, molecular imaging has attracted considerable interest among researchers in the imaging sciences because it enables quantification of biological activities at the cellular and molecular levels [27]. For example, changes in activities of the receptor tyrosine kinase EGFR can be visualized and quantified through the optical imaging of reconstituted luciferase [8]. PET enables detection and quantification of molecular processes such as glucose metabolism, angiogenesis, apoptosis, and necrosis [28, 29]. Radiolabeled annexin V uptake by apoptotic and necrotic cells is used to measure apoptosis, necrosis, and other disease processes using PET [30, 31]. Chelated gadolinium attached to small peptides recognizes cell receptors and quantify receptor activities using magnetic imaging techniques. Similarly, microbubbles and nanobubbles attached to antibodies such as anti-P-selectin may be used to image targeted molecules associated with inflammation, angiogenesis, intravascular thrombus, and tumors [32].
Despite the obvious advantages of imaging, there are many obstacles to the integration of biomedical images from different studies to conduct integrative analyses. First, there is significant heterogeneity and variability in imaging-based measurements. Unlike DNA sequence analysis or gene expression analysis whose data alphabets and fold change measurements are readily standardized, imaging is often viewed as being both a fundamentally subjective medium and significantly more complex due to the assay methods used. In particular, postprocessing, analysis, and interpretation are generally highly idiosyncratic to the particular study. Even apparently simple steps can have significant variations.
For example, imaging experts identify lesions and draw regions of interests (ROIs) around the lesions using image processing software. Previous studies have shown that there is significant variability across experts as well as across image processing algorithms implemented in different platforms [3, 4, 33]. There are many approaches to extract measurements from ROIs. For example, the SUV used in PET measures relative cellular uptake of an imaging probe. The most commonly used PET imaging probe in cancer diagnosis is [18F]-FDG that is used to measure glucose metabolisms. This single parameter has several variations: max SUV (SUVmax), mean SUV (SUVmean), and relative SUVs. SUVmax and SUVmean are absolute values whereas rSUV is a ratio of the SUV value of one anatomical region to that of another. The multiple ways of quantifying lesion SUV make it difficult to compare analyses across different studies [5].
As another motivating example, the rich data in biomedical images are not reused. The integration of imaging data from various experiments in order to promote new knowledge has not been effectively achieved. The biomedical imaging community requires a bioinformatics infrastructure that would enable researchers to search, access, and analyze this large amount of imaging data. One of the main challenges may be that the complexity and diversity of imaging data far exceeds genomic and proteomic data.
Despite these difficulties, significant efforts have been made to build imaging resources and databases. The National Biomedical Imaging Archive (NBIA) and The Cancer Imaging Archive developed by National Cancer Institute are large in vivo image repositories [34]. Images are publicly available to researchers in the biomedical research community for many purposes including lesion detection software development and the quantitative imaging assessment of drug responses [34]. Researchers are able to query and download images from 15 major imaging modalities and 18 anatomical sites for different tumor types. Early efforts to publish primary data along with analyzed results in the context of peer-reviewed journals have begun as well [35].
Another imaging-related database is the Molecular Imaging and Contrast Agent Database (MICAD) created by the National Center for Biotechnology Information [20]. Instead of images, the MICAD database focuses on molecular imaging agents and has 1,373 molecular imaging agents listed as of February 2013. In addition, there are thousands more in “pending” status. The imaging agents in MICAD encompass radioactive labeled small molecules [15], nanoparticles [16], proteins such as antibodies [36] and fluorescent tagged proteins [18], and labeled cells such as stem cells for tracking homing to tumors [19]. Moreover, there are 170 diverse biological applications corresponding to the imaging agents, ranging from biological transporter imaging, cell tracking, angiogenesis imaging to drug resistance and disease detection [20].
Because the emphasis of the MICAD database is on molecular imaging, many imaging agents have specific molecular targets. In total, there are unique 350 molecular targets, including mRNA, integrins, growth factors, insulin receptor, stem cells, etc. There are redundancies in these target annotations as a target can be used to annotate elsewhere using its synonyms. However, there is no formalized knowledge framework for this rich database that is publicly available.
Ontologies and their applications in biomedical research
An ontology is a framework that represents knowledge entities in a specific domain as well as relationships between entities [22]. The ontological structure is an ideal framework due to its ability to integrate heterogeneous and complex knowledge. It can be used to discover underlying associations in a knowledge domain.
It consists of three main components: terms/classes, properties/attributes, and instances. Terms describe important entities in a knowledge domain. Classes are structured in an IS-A hierarchy where each subclass is a more specific type of its superclass. Properties (or attributes) of each term describe the characteristics of classes. Instances are specific examples of terms in the ontology.
Also, by formally defining terms and synonyms of terms in a domain, an ontology facilitates the elimination of terminology variation and ambiguity, and thus can be used to link data and knowledge from different sources. For example, Gene Ontology is a major bioinformatics initiative that provides a controlled vocabulary for annotations of gene products [37]. It covers three areas: cellular component, molecular function, and biological process. Gene ontology (GO) is now widely used in annotations of gene expression [37]. For example, in microarray analysis, up- and downregulated genes are annotated using GO to represent common molecular functions or biological processes to identify the common theme among differentially regulated genes [38]. For example, enrichment of differentially expressed genes in a tumor reveals growth factor activity. In an area where data mining is tedious, ontologies allows for intelligent data mining.
Ontologies are reusable and can be linked with other ontologies for specific applications [39]. For example, the Foundational Model of Anatomy (FMA) is a domain ontology for correlating different views of anatomy. Washington et al. integrated FMA and other ontologies to associate human diseases with animal models [40].
Other widely used biomedical ontologies include NCI thesaurus (NCIt) and medical subject headings (MeSH) [41]. NCI thesaurus is a controlled terminology developed by the National Cancer Institute in collaboration with many partners, focusing on enabling the communication of information in cancer research. MeSH is the National Library of Medicine-controlled vocabulary thesaurus used for indexing articles in PubMed. Because of its comprehensiveness, it is used as a reference terminology in other applications [42].
Informatics in Imaging Biomarker Research
RadLex (https://www.rsna.org/RadLex.aspx) is a unified language of radiology terms for standardized indexing and retrieval of radiology information resources [43]. With more than 30,000 terms, RadLex satisfies the needs of software developers, system vendors, and radiology users by adopting the best features of existing terminology systems while producing new terms to fill critical gaps. For example, researchers and clinicians can use the RadLex terminology to annotate radiological images [44]. It unifies and supplements other lexicons and standards, such as SNOMED-CT and DICOM.
Annotation and Image Markup (AIM) is an information model focused on clinical imaging that provides human observers with the ability to record explanatory or descriptive information about their observations and to attach this information to specific locations within an image [45]. Using RadLex terms, it provides a standardized information model compatible with DICOM-SR, XML, and HL7 formats for such observations.
Formal Definition as Basis for Validation and Qualification Framework
The lack of consensus methods and carefully characterized performance impedes the widespread availability of urgently needed quantitative imaging techniques in medicine. A precondition for use is the demonstration of performance according to recognized descriptive statistics computed in a defined patient population with a specific biological phenomenon associated with a known disease state, supported by evidence in large patient populations, and externally validated. Not yet merged into a framework that supports the collaborative work needed to meet the potential of quantitative imaging analysis are the application of advanced statistical techniques, the development of controlled vocabularies and service-oriented architecture for processing large image archives. With the availability of tools for the automatic ontology-based annotation of datasets with terms from biomedical ontologies such as those made possible by the Quantitative Imaging Biomarker Ontology (QIBO), coupled with imaging archives and a means for the batch selection and processing of imaging and clinical data, we believe that imaging will go through a similar increase in capability analogous to the gains advanced sequencing techniques have brought to molecular biology.
To address the need for informatics methods in imaging research, validation, and qualification, the goal of this project is to develop a structured knowledge representation using an ontology to integrate heterogeneous knowledge in imaging biomarkers. The domain of our ontology is defined as imaging biomarkers for both preclinical and clinical applications.
Materials and Methods
We have created the QIBO to provide a basis for standardizing semantics inclusive of the terms as well as the relationships among them.
Initial Curation to Collect Terms
To gather terms, relationships, and properties of the terms in the imaging biomarker ontology, we started with a literature review of the journal Molecular Imaging and Biology. We sampled 22 articles published since 2006. The inclusion criteria was that articles made nontrivial use of quantitative imaging measurements, which was not true of many chemistry/probe development papers. Articles were randomly sampled and added until it was subjectively determined that the scope of the concepts had begun to elucidate enough structure such that an ontology could be designed around it. This work was by two individuals with 23 years of imaging research experience in total with all work reviewed by both and the more senior arbitrating conflicts. Further, we also distilled terms associated with 95 putative biomarkers in oncology indications from 43 articles in such journals as JNM, JMRI, Cell, Molecular Imaging and Biology, and Transgenic Research; 47 putative biomarkers in cardiovascular indications from 30 articles in such journals as JNM, Molecular Therapy, Stroke, and Circulation; 52 putative biomarkers in neurology indications from 25 journals such as Nuclear Medicine and Biology, NeuroImage, and Nuclear Instruments & Methods in Physics Research; 22 putative biomarkers in musculoskeletal indications from 6 articles in such journals as EJNM and Molecular Imaging; 5 putative biomarkers in endocrinology indications from 3 articles in Proceedings of the National Academy of Sciences and Vanderbilt University Institute of Imaging Science conference proceedings; and 4 putative biomarkers from 19 articles in pulmonary indications from 19 articles in such journals as European Respiratory Journal, Proceedings of American Thoracic Society, Medical Physics, and Thoracic Imaging. We examined the methods and results of experiments reported in the papers, focusing on abstracting terms and relationships that characterize and annotate an imaging biomarker. This approach allows us to examine specific instances of biomarkers and to be able to build the ontology from real world examples.
Reusing Other Publicly Available Ontologies
To build the imaging biomarker ontology, we reviewed a range of publicly available ontologies and terminologies, including MeSH [41], NCI thesaurus [46], GO [37], FMA [39], Disease Ontology [47], and BIRNLex (a controlled terminology for Biomedical Informatics Research Network) [48] to adopt or adapt them wherever possible. For example, entities of anatomical structure were imported from FMA and entities of biological process were extracted from GO, MeSH, and NCI thesaurus.
Our solution to integrating multiple ontologies is to minimally and unambiguously reference the external class from within QIBO. This is achieved by specifying the external source ontology, the external source class from the source ontology and the QIBO class that references the external source class. In our case, most of the ontologies we reuse in QIBO are fairly stable. We coded the minimal references directly in the ontology as annotation properties in Web Ontology Language (OWL).
Design Decisions
The domain of the ontology includes imaging biomarkers for both preclinical and clinical applications featuring molecular imaging because of its richness and biological specificity.
We have built the OWL-based ontology using Protégé, a widely used and freely available ontology editing tool [49]. Protégé supports two ontology modeling paradigms: OWL and frames. Despite many similarities between the two languages, OWL provides description logic reasoning capability with high expressive power [50]. Classes can be asserted directly in the ontology in both OWL and frames, describing necessary conditions. Only necessary and sufficient conditions can be defined in OWL to specify new classes where an OWL classifier can be run to generate inferred hierarchy. Inferred hierarchy is particularly suitable for representing the heterogeneous knowledge in imaging biomarkers. It allows the defining of newly discovered biomarkers. OWL enables powerful knowledge reasoning. It is capable of conveying complexity and richness in imaging biomarker research.
Based on our curation efforts, we first identified top-level terms that capture major entities appearing in an imaging biomarker experiment. We also created synonyms and definitions for classes in the ontology. To ensure consistency, Is–A relationship (subsumption) is strictly conserved in every branch of the ontology. Other relationships that organize the terms in a structured and meaningful way are defined as properties, such as the relationships between the first level classes.
We anticipate that QIBO will be extended to relate and link to other established ontologies such as FMA [51], GO [52], SNOMED [53], and RadLex [54] as well as other standardized domain ontologies such as from the Open Biological and Biomedical Ontologies Foundry [55], leveraging the Basic Formal Ontology (BFO) [56] upper ontology for alignment through a shared abstract level. It also incorporates NBIA [57] and AIM UML models, associated common data elements and underlying NCIt and other ontology concepts.
Results
Upper Level Terms and Their Relations
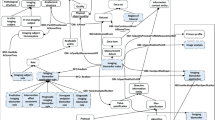
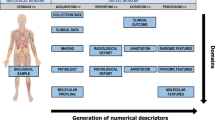
We first identified top-level terms of QIBO in the temporal order in which they would appear in an imaging biomarker experiment (Fig. 1). We then formally defined the first level classes and their semantic relationships (Fig. 2). The ontology contains 567 classes, of which IMAGING INSTRUMENT and BIOLOGICAL TARGET are the two largest classes in terms of the number of subconcepts (Figs. 3 and 4). Term names in the following description are denoted in CAPS for clarity.
Distribution of classes in each of the top-level terms in the Quantitative Imaging Biomarker Ontology: postprocessing algorithm (20), biological intervention (24 concepts), biomarker use (32 concepts), imaging agent (57 concepts), biological subject (57 concepts), quantitative imaging biomarker (57 concepts), indicated biology (92 concepts), biological target (113 concepts), and imaging instrument (115 concepts)
We also created synonyms and definitions for classes in the ontology. To ensure consistency, Is–A relationship (subsumption) is strictly conserved in every branch of the ontology (Fig. 5). Other relationships are defined as properties, such as relationships between the first level classes (Fig. 2).
BIOLOGICAL SUBJECT refers to the living organism, or part of a living organism, on which an experimental or clinical imaging study is performed. We created 56 subclasses under this branch. BIOLOGICAL INTERVENTION refers to a procedure performed on the BIOLOGICAL SUBJECT either to perturb the biology (e.g., conditional gene expression) or for therapeutic purposes (e.g., drug administration; Fig. 2). There are in total 24 concepts in BIOLOGICAL INTERVENTION.
The IMAGING BIOMARKER MEASUREMENT is a central term in the ontology and has relationships to many of the other upper classes. IMAGING BIOMARKER MEASUREMENT has subclasses, ANATOMICAL MEASUREMENT (e.g., length and volume) and FUNCTIONAL MEASUREMENT (e.g., metabolic rate biomarker and permeability biomarker), which are the super classes for 28 and 18 concepts, respectively.
IMAGING INSTRUMENT, as the largest branch with 115 concepts in the ontology, includes conventional anatomical imaging instruments (e.g., CT and MRI) as well as molecular and functional imaging instruments (e.g., microscope and bioluminescence imaging). ALGORITHM describes image processing, analytic techniques and/or modeling methods used to extract the IMAGING BIOMARKER MEASUREMENT. There are 20 different algorithm concepts in the branch. Although this term often is not mentioned with the IMAGING BIOMARKER MEASUREMENT, it is an indispensable step in the acquisition of the measurement. IMAGING AGENT is an exogenous material (e.g., contrast agent or molecular probe) optionally administered to the BIOLOGICAL SUBJECT that is used to visualize some component or process within the subject. IMAGING AGENT contains 57 subclasses.
BIOLOGICAL TARGET, as the second largest branch with 113 concepts in the ontology, is the part of the biological subject that is optionally targeted by the IMAGING AGENT; it is visualized in the image and measured in order to create the imaging biomarker. BIOLOGICAL TARGET refers to biological components such as biomolecules, cells, or anatomical structure/space. The subclasses are populated using terms from an existing ontology NCI thesaurus, as well as our own terms. For example, subclasses of enzyme in molecular target are shared with those of enzyme in NCI thesaurus; multicellular organ level target shares subclasses with skeletal system part in NCI thesaurus. INDICATED BIOLOGY includes 55 biological process and 37 disease concepts, which are populated using terms from GO, MeSH, and Disease Ontology. For example, terms under DISEASE reference equivalent classes in the Disease Ontology via DOID using the annotation property (Fig. 6). There are 24 of the 37 disease concepts annotated with DOIDs. Similarly, BIOLOGICAL PROCESS is semantically equivalent to the same term in one of the three branches of GO, and thus 46 of the 55 concepts in Biological Process are shared with the GO biological_process branch. Although the biological process branch in GO is comprehensive in terms of molecular and cellular processes, we also include disease level processes with ill-defined genetics in order to better cover clinical applications of imaging biomarkers, such concepts as thrombosis and metastasis. INDICATED BIOLOGY and BIOLOGICAL TARGET can both be considered as functional annotations of the IMAGING BIOMARKER MEASUREMENT. We make such a distinction in order to conserve the subsumption property of the ontology. INDICATED BIOLOGY and BIOLOGICAL TARGET are two classes that overlap significantly with other ontologies.
BIOMARKER USE is the application of the imaging biomarker, whether for drug development, for a biological discovery such as signaling pathway identification, or for clinical practice in diagnosis, screening, staging, etc. This branch consists of 32 concepts.
Relationships Between Upper Level Terms
Figure 2 shows relationships between upper level terms. To highlight QUANTITATIVE IMAGING BIOMARKER as the central term in the ontology, we show the relationships from the central term to the other terms in Fig. 2. For every relationship, there is an inverse relationship in the reversed direction (not shown). For example, QUANTITATIVE IMAGING BIOMARKER is_a_measure of INDICATED BIOLOGY, and INDICATED BIOLOGY is_quantified_by the QUANTITATIVE IMAGING BIOMARKER. Relationships are denoted in italics for clarity. The representation of relationships between upper level terms not only elucidates semantic associations between the terms, but also allows inferences from one term to another.
In conforming to ontological hierarchy, the relations between top-level concepts, like other attributes of concepts, are inherited by their subclasses. In another word, the relations between top-level concepts are propagated to subclasses in their respective subclasses. Thus, the relationship between two concepts from different main branches is dictated by the relationship between their top-level concepts if these two concepts are two of the nine main branches. The relationship between any two concepts within a main branch strictly follows the subsumption property.
Applications
-
1.
Discovery of Novel Biomarkers. Among the benefits of our approach is the ability to discover how biomarkers are related. For example, two biomarkers might be used for the same disease, but such relations cannot be represented by a flat list. The ontological structure in QIBO—both the hierarchical development of concepts, as well as the free graph-oriented relations among them—addresses these issues. QIBO enables semantically identical terms to be considered as one concept in order to enable precise semantics. This capability can be used for the discovery of novel biomarkers. We have identified an example of this use case in our curation (Fig. 7). Figure 7 shows two associations of measurement–target–disease.
-
the first association was identified in the ninth curated paper, illustrating that max photons per second per square centimeter per steradian (biomarker measurement 1) is a measure of cardiac muscle cells (target 1) for acute donor cell death (disease or pathological condition 1) [58].
-
the second association was identified in the 29th curated paper, which links myocardial blood flow measured as MBFp or milliliter per minute per milliliter in PET (biomarker measurement 2) with myocardium (target 2) and chronic ischemic heart disease (disease 2; Fig. 7a) [59].
As the targets are identical in the two associations, the two associations can be linked (Fig. 7b). Thus, biomarker measurement 1 is a potential new biomarker for disease 2, and similarly for biomarker measurement 2 and disease 1. Furthermore, QIBO can be mapped with other public ontologies to establish associations of biomarkers and other biological entities for new biological discovery.
-
2.
Validation and Qualification Framework. Enabled by the work in this paper, we are in the process of developing QI-Bench© (www.qi-bench.org), an open-source informatics tool which clinicians and researchers with diverse scientific backgrounds, users without informatics background, can use to characterize the performance of quantitative medical imaging to specify and support experimental activities for statistical validation using the QIBO and other resources accessible through the National Center for Biomedical Ontology’s BioPortal [60]. QI-Bench uses QIBO to guide the creation of an imaging biomarker knowledgebase with instances of the QIBO terms analogous in some ways to how Bio2RDF and other applications use GO, and we anticipate further parallels as development continues.
Discussion
This work presented here has several limitations. First, upper ontologies such as BFO can provide clear philosophical distinctions between things such as continuants and occurrents and can enable ontological alignment. Currently, QIBO does not derive from a standardized upper ontology because of the obfuscation of the specific terms that are of actual use to imaging researchers. Second, reusing other ontologies avoids duplicating efforts and allows for integration between different ontologies as well as interoperability of applications using them. However, the actual implementation of seamlessly merging existing ontologies into a developing ontology is still an unsolved challenge and our approach may not be ideal. Third, since ontology development is a continual process, a more formalized structure must be developed for the ongoing development and maintenance of the ontology by a larger community. For instance, the addition of concepts and level of detail of relations should be arbitrated in a fair and consistent manner and unintentional duplication must be avoided. QIBO is still in its early stages but its structure, content, and documentation follow and is evaluated against terminology review principles [61].
The Quantitative Imaging Biomarker Ontology we have developed integrates knowledge from multiple fields, spanning the context for use, and assay methods. Context for use is represented by biology of interest (biological target, indicated biology, and biomarker application). The ontology also includes both clinical terms (e.g., DISEASE) and terms used in basic biological research (e.g., BIOLOGICAL PROCESS). Assay methods are represented by upper classes, including image technique (e.g., IMAGING AGENT and IMAGING INSTRUMENT), and method of quantitation (e.g., IMAGE PROCESSING and ANALYSIS ALGORITHM).
Integration of these different fields will facilitate biomarker discovery and accelerate the translation of imaging biomarkers from research to clinical use. In addition, we reuse existing ontologies whenever possible. The complete QIBO is available to the public on the BioPortal website [62].
The subsumption (IS–A) relationship and other types of relationships between terms maintained in the ontology enable semantically meaningful retrievals of related terms in QIBO. For example, the ontology may be used to annotate instances of imaging biomarkers. Similar biomarker instances can be retrieved based on their semantic similarity.
To our knowledge, this is the first ontological representation of knowledge in imaging biomarker research. Tulipano et al. developed a terminology in molecular imaging, focusing on bridging the imaging domain with gene product function, process and location described in GO [63, 64]. However, the top level terms in this ontology only encompass imaging (IMAGING INSTRUMENT, AMPLIFICATION TECHNIQUE, IMAGING PROBE), BIOLOGICAL TARGET (IMAGING TARGET), and MOLECULARE IMAGING ENTITY, which is too broad to specify and differentiate different imaging biomarkers.
RadLex and QIBO are two closely related knowledge representations in imaging, but each with a different focus. The RadLex annotations are semantic descriptors of qualitative interpretation on images, whereas QIBO semantically captures quantitative features that are computationally derived.
One promising use of the ontology is to bridge the QIBO terms according to their relationships and create statements represented as RDF triples and store them in an RDF store such as Bio2RDF. We have begun this work as one aspect of QI-Bench. By doing so, we are building semantically rich specifications for linked data. This representation will provide a direct benefit in allowing integrated knowledge across imaging and non-imaging data sets, as well as enabling applications to assemble/transform the set of RDF triples to SPARQL queries.
Data integration on the conceptual level enabled by this method abstracts out implementation details to increase the accessibility of data. Data integration could be performed across the genomic, gene expression, clinical phenotype, and imaging data, using federated SPARQL queries and inferencing to formulate testable hypotheses and associated datasets for the validation of a new imaging biomarker based on a linked data specification of the biomarker using terms from QIBO.
Conclusions
We have developed QIBO to support imaging biomarker research. It integrates heterogeneous knowledge in the field of quantitative imaging and bridges preclinical and clinical imaging biomarker research. Presently, we validated the ontology using published imaging biomarker data. We have demonstrated its utility in various applications associated with the QI-Bench program such as data retrieval, mining new information from the literature and discovering novel imaging biomarkers.
References
Fuchs VR, Sox Jr, HC: Physicians' views of the relative importance of thirty medical innovations. Health Aff (Millwood) 20(5):30–42, 2001
Atkinson AJ, et al: Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clinical pharmacology and therapeutics 69(3):89–95, 2001
Zhao B, James LP, Moskowitz CS, Guo P, et al: Evaluating variability in tumor measurements from same-day repeat CT scans of patients with non-small cell lung cancer. Radiology 252(1):263–272, 2009
Sheikh HR, Sabir MF, Bovik AC: A statistical evaluation of recent full reference image quality assessment algorithms. IEEE Transactions on Image Processing: a Publication of the IEEE Signal Processing Society 15(11):3440–3451, 2006
Buckler AJ, Boellaard R: Standardization of quantitative imaging: the time is right, and 18F-FDG PET/CT is a good place to start. Journal of Nuclear Medicine: Official Publication, Society of Nuclear Medicine 52(2):171–172, 2011
Wong D: Liaison Committee Discusses Possible Radiotracer Sharing Clearinghouse, in American College of Neuropsychopharmacology, 2006
Wong DF: Imaging in drug discovery, preclinical, and early clinical development. Journal of Nuclear Medicine: Official Publication, Society of Nuclear Medicine 49(6):26N–28N, 2008
Gene Ontology Consortium: Creating the gene ontology resource: design and implementation. Genome Res 11(8):1425–1433, 2001
Ashburner M, Ball CA, Blake JA, Botstein D, et al: Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25(1):25–29, 2000
Romero-Zaliz RC, Rubio-Escudero C, Cobb JP, Herrera F, Cordon O, Zwir I: A multiobjective evolutionary conceptual clustering methodology for gene annotation within structural databases: a case of study on the Gene Ontology Database. IEEE Transactions on Evolutionary Computation 12(6):679–701, 2008
Brazma A, Hingamp P, Quackenbush J, Sherlock G, et al: Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 29(4):365–371, 2001
Sirota M, Dudley JT, Kim J, Chiang AP, Morgan AA, Sweet-Cordero A, Sage J, Butte AJ: Discovery and preclinical validation of drug indications using compendia of public gene expression data. Science translational medicine 3(96):96ra77, 2011
Brown MS, Shah SK, Pais RC, Lee YZ, McNitt-Gray MF, Goldin JG, Cardenas AF, Aberle DR: Database design and implementation for quantitative image analysis research. IEEE Trans Inf Technol Biomed 9(1):99–108, 2005
Maier D, Kalus W, Wolff M, Kalko SG, et al: Knowledge management for systems biology a general and visually driven framework applied to translational medicine. BMC Syst Biol 5:38, 2011
Toyohara J, Kumata K, Fukushi K, Irie T, Suzuki K: Evaluation of 4'-[methyl-14C]thiothymidine for in vivo DNA synthesis imaging. Journal of Nuclear Medicine: Official Publication, Society of Nuclear Medicine 47(10):1717–1722, 2006
Yuk SH, Oh KS, Cho SH, Lee BS, Kim SY, Kwak BK, Kim K, Kwon IC: Glycol chitosan/heparin immobilized iron oxide nanoparticles with a tumor-targeting characteristic for magnetic resonance imaging. Biomacromolecules 12(6):2335–2343, 2011
Veenendaal LM, Jin H, Ran S, Cheung L, Navone N, Marks JW, Waltenberger J, Thorpe P, Rosenblum MG: In vitro and in vivo studies of a VEGF121/rGelonin chimeric fusion toxin targeting the neovasculature of solid tumors. Proc Natl Acad Sci U S A 99(12):7866–7871, 2002
Wen X, Lyu MA, Zhang R, Lu W, Huang Q, Liang D, Rosenblum MG, Li C: Biodistribution, pharmacokinetics, and nuclear imaging studies of 111In-labeled rGel/BLyS fusion toxin in SCID mice bearing B cell lymphoma. Molecular Imaging and Biology: MIB: the Official Publication of the Academy of Molecular Imaging 13(4):721–729, 2011
Wang HH, Wang YX, Leung KC, Au DW, et al: Durable mesenchymal stem cell labelling by using polyhedral superparamagnetic iron oxide nanoparticles. Chemistry 15(45):12417–12425, 2009
http://www.ncbi.nlm.nih.gov/books/NBK5330/, Molecular Imaging and Contrast Agent Database (MICAD). 2011
Quon A, Gambhir SS: FDG-PET and beyond: molecular breast cancer imaging. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology 23(8):1664–1673, 2005
Noy NFM, DL: Ontology development 101: a guide to creating your first ontology. Stanford Knowledge Systems Laboratory technical report KSL-01-05 and Stanford Medical Informatics technical report SMI-2001-0880, 2001
Jaffer FA, Weissleder R: Molecular imaging in the clinical arena. JAMA: The Journal of the American Medical Association 293(7):855–862, 2005
Buckler AJ, Bresolin L, Dunnick NR, Sullivan DC: A collaborative enterprise for multi-stakeholder participation in the advancement of quantitative imaging. Radiology 258(3):906–914, 2011
Buckler AJ, Bresolin L, Dunnick NR, Sullivan DC, et al: Quantitative imaging test approval and biomarker qualification: interrelated but distinct activities. Radiology 259(3):875–884, 2011
Smith JJ, Sorensen AG, Thrall JH: Biomarkers in imaging: realizing radiology's future. Radiology 227(3):633–638, 2003
Li W, Li F, Huang Q, Frederick B, Bao S, Li CY: Noninvasive imaging and quantification of epidermal growth factor receptor kinase activation in vivo. Cancer research 68(13):4990–4997, 2008
West C, Charnley N: The potential of PET to increase understanding of the biological basis of tumour and normal tissue response to radiotherapy. Br J Radiol November 2005 Supplement_28:50–54; doi:10.1259/bjr/83746792, 2005
Rudin M, Weissleder R: Molecular imaging in drug discovery and development. Nat Rev Drug Discov 2(2):123–131, 2003
Toretsky J, Levenson A, Weinberg IN, Tait JF, Uren A, Mease RC: Preparation of F-18 labeled annexin V: a potential PET radiopharmaceutical for imaging cell death. Nuclear medicine and biology 31(6):747–752, 2004
Zijlstra S, Gunawan J, Burchert W: Synthesis and evaluation of a 18F-labelled recombinant annexin-V derivative, for identification and quantification of apoptotic cells with PET. Applied Radiation and Isotopes: Including data, Instrumentation and Methods for use in agriculture, industry and medicine 58(2):201–207, 2003
Klibanov AL: Ligand-carrying gas-filled microbubbles: ultrasound contrast agents for targeted molecular imaging. Bioconjugate Chemistry 16(1):9–17, 2005
Habte F, B.S., Keren S, Doyle TC, Levin CS, Paik DS: In situ study of the impact of inter- and intra-reader variability on region of interest (ROI) analysis in preclinical molecular imaging. American Journal of Nuclear Medicine and Molecular Imaging, 2013 (in press)
Archive NBI: https://cabig.nci.nih.gov/tools/NCIA, 2011
Buckler AJ SL, Petrick N, McNitt-Gray M, Zhao B, Fenimore C, Reeves AP, Mozley PD, Avila RS: Data sets for the qualification of CT as a quantitative imaging biomarker in lung cancer. Optics express 18(14):16, 2010
Veenendaal LM, Jin H, Ran S, Cheung L, Navone N, Marks JW, Waltenberger J, Thorpe P, Rosenblum MG: In vitro and in vivo studies of a VEGF121/rGelonin chimeric fusion toxin targeting the neovasculature of solid tumors. Proceedings of the National Academy of Sciences of the United States of America 99(12):7866–7871, 2002
Ashburner M, Ball CA, Blake JA, Botstein D, et al: Gene ontology: tool for the unification of biology, The Gene Ontology Consortium. Nature genetics 25(1):25–29, 2000
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, et al: Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102(43):15545–15550, 2005
Rosse C, Mejino Jr, JL: A reference ontology for biomedical informatics: the Foundational Model of Anatomy. Journal of biomedical informatics 36(6):478–500, 2003
Washington NL, Haendel MA, Mungall CJ, Ashburner M, Westerfield M, Lewis SE: Linking human diseases to animal models using ontology-based phenotype annotation. PLoS biology 7(11):e1000247, 2009
Lowe HJ, Barnett GO: Understanding and using the medical subject headings (MeSH) vocabulary to perform literature searches. JAMA: The Journal of the American Medical Association 271(14):1103–1108, 1994
Swanson DR: N.R.S.a.V.I.T., Ranking indirect connections in literature-based discovery: The role of Medical Subject Headings (MeSH). Journal of the American Society for Information Science and Technology 57(11):1427–1439, 2006
Rubin DL: Creating and curating a terminology for radiology: ontology modeling and analysis. Journal of Digital Imaging: the Official Journal of the Society for Computer Applications in Radiology 21(4):355–362, 2008
Rubin DL, Rodriguez C, Shah P, Beaulieu C: iPad: semantic annotation and markup of radiological images. AMIA. Annual Symposium proceedings/AMIA Symposium. AMIA Symposium, 2008, pp 626–30
Rubin DL, M.P., Kleper V, Supekar K, Channin DS: Medical imaging on the semantic web: annotation and image Markup, in AAAI Spring Symposium Series, Semantic Scientific Knowledge Integration 2008: Stanford University
Thesaurus N: https://cabig.nci.nih.gov/tools/NCI_Thesaurus, 2011
Schriml LM, Arze C, Nadendla S, Chang YW, Mazaitis M, Felix V, Feng G, Kibbe WA: Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res, 2011. doi:10.1093/nar/gkr972
Bug WJ, Ascoli GA, Grethe JS, Gupta A, et al: The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics 6(3):175–194, 2008
Rubin DL, Noy NF, Musen MA: Protege: a tool for managing and using terminology in radiology applications. Journal of Digital Imaging: the Official Journal of the Society for Computer Applications in Radiology 20(Suppl 1):34–46, 2007
Wang H, Rector A, Drummond N, Horridge M. et al: Frames and OWL Side by Side. 9th International Protégé Conference, Stanford, California, USA, 2006
The Foundational Model of Anatomy ontology (FMA). Available from: http://sig.biostr.washington.edu/projects/fm/AboutFM.html. Accessed 27 November 2011
The Gene Ontology project. Available from: http://www.geneontology.org/, accessed 27 November 2011
SNOMED Clinical Terms® (SNOMED CT®). Available from: http://www.nlm.nih.gov/research/umls/Snomed/snomed_main.html. Accessed 27 November 2011
RSNA RadLex. Available from: http://www.rsna.org/informatics/radlex.cfm. Accessed 27 November 2011
The Open Biological and Biomedical Ontologies. Available from: http://www.obofoundry.org/. Accessed 27 November 2011
BFO Basic Formal Ontology. Available from: http://www.ifomis.org/bfo. Accessed 27 November 2011
National Biomedical Imaging Archive. 2011; Available from: https://cabig.nci.nih.gov/tools/NCIA. Accessed 27 November 2011
Gheysens O, Lin S, Cao F, Wang D, Chen IY, Rodriguez-Porcel M, Min JJ, Gambhir SS, Wu JC: Noninvasive evaluation of immunosuppressive drug efficacy on acute donor cell survival. Molecular imaging and biology: MIB: the official publication of the Academy of Molecular Imaging 8(3):163–170, 2006
Knaapen P, Bondarenko O, Beek AM, Gotte MJ, et al: Impact of scar on water-perfusable tissue index in chronic ischemic heart disease: Evaluation with PET and contrast-enhanced MRI. Molecular imaging and biology: MIB: the official publication of the Academy of Molecular Imaging 8(4):245–251, 2006
Bench Q: (www.qi-bench.org)
Cimino JJ, Hayamizu TF, Bodenreider O, Davis B, Stafford GA, Ringwald M: The caBIG terminology review process. Journal of biomedical informatics 42(3):571–580, 2009
Bioportal Qo: http://bioportal.bioontology.org/ontologies/46348?p=terms
Tulipano PK, Millar WS, Cimino JJ: Linking molecular imaging terminology to the gene ontology (GO). Pac Symp Biocomput p. 613–23, 2003
Tulipano PK, Tao Y, Millar WS, Zanzonico P, et al: Natural language processing and visualization in the molecular imaging domain. Journal of biomedical informatics 40(3):270–281, 2007
Acknowledgments
These activities have been supported in part with federal funds from the National Institute of Standards and Technology, Department of Commerce, under Cooperative Agreement No. 70NANB10H223 (which partially funds the QI-Bench program). We would also like to acknowledge Yi Liu of the Biomedical Informatics Program at Stanford University annotating linked terms in QIBO shared with Disease Ontology.
Author information
Authors and Affiliations
Corresponding author
Additional information
Andrew J. Buckler and Tiffany Ting Liu contributed equally to this study.
Rights and permissions
About this article
Cite this article
Buckler, A.J., Ouellette, M., Danagoulian, J. et al. Quantitative Imaging Biomarker Ontology (QIBO) for Knowledge Representation of Biomedical Imaging Biomarkers. J Digit Imaging 26, 630–641 (2013). https://doi.org/10.1007/s10278-013-9599-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-013-9599-2