Abstract
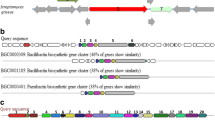
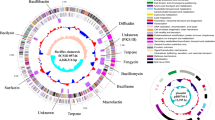
Genomic studies provide deeper insights into secondary metabolites produced by diverse bacterial communities, residing in various environmental niches. This study aims to understand the potential of a biosurfactant producing Bacillus sp. AM13, isolated from soil. An integrated approach of genomic and chemical analysis was employed to characterize the antibacterial lipopeptide produced by the strain AM13. Genome analysis revealed that strain AM13 harbors a nonribosomal peptide synthetase (NRPS) cluster; highly similar with known biosynthetic gene clusters from surfactin family: lichenysin (85 %) and surfactin (78 %). These findings were substantiated with supplementary experiments of oil displacement assay and surface tension measurements, confirming the biosurfactant production. Further investigation using LCMS approach exhibited similarity of the biomolecule with biosurfactants of the surfactin family. Our consolidated effort of functional genomics provided chemical as well as genetic leads for understanding the biochemical characteristics of the bioactive compound.





Similar content being viewed by others
References
Al-Bahry SN, Al-Wahaibi YM, Elshafie AE, Al-Bemani AS, Joshi SJ, Al-Makhmari HS, Al-Sulaimani HS (2013) Biosurfactant production by Bacillus subtilis B20 using date molasses and its possible application in enhanced oil recovery. Int Biodeterior Biodegrad 81:141–146. doi:10.1016/j.ibiod.2012.01.006
Aleti G, Sessitsch A, Brader G (2015) Genome mining: prediction of lipopeptides and polyketides from bacillus and related firmicutes. Comput Struct Biotechnol J 13:192–203. doi:10.1016/j.csbj.2015.03.003
Angiuoli SV, Gussman A, Klimke W, Cochrane G, Field D, Garrity G, Kodira CD, Kyrpides N, Madupu R, Markowitz V, Tatusova T, Thomson N, White O (2008) Toward an online repository of Standard Operating Procedures (SOPs) for (meta)genomic annotation. Omi A J Integr Biol 12:137–141. doi:10.1089/omi.2008.0017
Ansari MZ, Yadav G, Gokhale RS, Mohanty D (2004) NRPS-PKS: a knowledge-based resource for analysis of NRPS-PKS megasynthases. Nucleic Acids Res 32:405–413. doi:10.1093/nar/gkh359
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F et al (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi:10.1186/1471-2164-9-75
Ben Ayed H, Jridi M, Maalej H, Nasri M, Hmidet N (2014) Characterization and stability of biosurfactant produced by bacillus mojavensis A21 and its application in enhancing solubility of hydrocarbon. J Chem Technol Biotechnol 89:1007–1014. doi:10.1002/jctb.4192
Bergmann S, Schümann J, Scherlach K, Lange C, Brakhage AA, Hertweck C (2007) Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol 3:213–217. doi:10.1038/nchembio869
Branquinho R, Sousa C, Lopes J, Pintado ME, Peixe LV, Osorio (2014) Differentiation of Bacillus pumilus and Bacillus safensis using MALDI-TOF-MS. PLoS One 9:e116426. doi:10.1371/journal.pone.0110127
Carrillo PG, Mardaraz C, Pitta-Alvarez SI, Giulietti AM (1996) Isolation and selection of biosurfactant-producing bacteria. World J Microbiol Biotechnol 12:82–4. doi:10.1007/BF00327807
Chevreux B, Wetter T, Suhai S (1999) Genome sequence assembly using trace signals and additional sequence information. Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB) 99:45–56
Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57:2259–2261. doi:10.1099/ijs.0.64915-0
Dhillon BK, Laird MR, Shay JA, Winsor GL, Lo R, Nizam F, Pereira SK, Waglechner N, McArthur AG, Langille MG, Brinkman FS (2015) IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res 43:W104–8. doi:10.1093/nar/gkv401
Domingos DF, de Faria AF, de Souza GR, Eberlin MN, Greenfield P, Zucchi TD, Melo IS, Tran-Dinh N, Midgley D, de Oliveira VM (2015) Genomic and chemical insights into biosurfactant production by the mangrove-derived strain Bacillus safensis CCMA-560. Appl Microbiol Biotechnol 99:3155–3167. doi:10.1007/s00253-015-6377-8
Donio MBS, Ronica FA, Viji VT, Velmurugan S, Jenifer JS, Michaelbabu M, Dhar P, Citarasu T (2013) Halomonas sp. BS4, a biosurfactant producing halophilic bacterium isolated from solar salt works in India and their biomedical importance. Springerplus 2:149. doi:10.1186/2193-1801-2-149
Doroghazi JR, Albright JC, Goering AW, Ju KS, Hainess RR, Tchalukov KA, Labeda DP, Kelleher NL, Metcalf WW (2014) A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol 10:963–8. doi:10.1038/nchembio.1659
Grant JR, Stothard P (2008) The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:181–184. doi:10.1093/nar/gkn179
Harvey AL, Edrada-Ebel R, Quinn RJ (2015) The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov 14:111–129. doi:10.1038/nrd4510
Heerklotz H, Seelig J (2007) Leakage and lysis of lipid membranes induced by the lipopeptide surfactin. Eur Biophys J 36:305–314. doi:10.1007/s00249-006-0091-5
Kapley A, Tanksale H, Sagarkar S, Prasad AR, Kumar RA, Sharma N, Qureshi A, Purohit HJ (2016) Antimicrobial activity of Alcaligenes sp. HPC 1271 against multidrug resistant bacteria. Funct Integr Genomics 16:57–65. doi:10.1007/s10142-015-0466-8
Konz D, Doekel S, Marahiel M a (1999) Molecular and biochemical characterization of the protein template controlling biosynthesis of the lipopeptide lichenysin molecular and biochemical characterization of the protein template controlling biosynthesis of the lipopeptide lichenysin. J Bacteriol 181:133–140, doi: 0021-9193/99/$04.0010
Koumoutsi A, Chen X, Henne A, Lieseqang H, Hitzeroth G, Franke P, Vater J, Borriss R (2004) Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol 186:1084–1096. doi:10.1128/JB.186.4.1084
Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi:10.1093/nar/gkm160
Lam KS (2007) New aspects of natural products in drug discovery. Trends Microbiol 15:279–289. doi:10.1016/j.tim.2007.04.001
Land M, Hauser L, Jun SR, Nookaew I, Leuze MR, Ahn TH, Karpinets T, Lund O, Kora G, Wassenaar T, Poudel S, Ussery DW (2015) Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics 15:141–61. doi:10.1007/s10142-015-0433-4
Lavermicocca P, Valerio F, Evidente A, Lazzaroni S, Corsetti A, Gobbetti M (2000) Purification and characterization of novel antifungal compounds from the sourdough lactobacillus plantarum strain 21B. Appl Environ Microbiol 66:4084–4090. doi:10.1128/AEM.66.9.4084-4090.2000
Liu Y, Lai Q, Dong C, Sun F, Wang L, Li G, Shao Z (2013) Phylogenetic diversity of the Bacillus pumilus group and the marine ecotype revealed by multilocus sequence analysis. PLoS One 8:1–11. doi:10.1371/journal.pone.0080097
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi:10.1093/nar/25.5.955
Lu L, Wang J, Xu Y, Wang K, Hu Y, Tian R, Yang B, Lai Q, Li Y, Zhang W, Shao Z, Lam H, Qian PY (2014) A high-resolution LC-MS-based secondary metabolite fingerprint database of marine bacteria. Sci Rep 4:6537. doi:10.1038/srep06537
Morikawa M, Hirata Y (2000) A study on the structure-function relationship of lipopeptide biosurfactants. 1488:211–218. doi:10.1016/S1388-1981(00)00124-4
Mukherjee S, Das P, Sivapathasekaran C, Sen R (2009) Antimicrobial biosurfactants from marine bacillus circulans: extracellular synthesis and purification. Lett Appl Microbiol 48:281–288. doi:10.1111/j.1472-765X.2008.02485.x
Naruse N, Tenmyo O, Kobaru S, Kamei H, Miyaki T, Konishi M, Oki T (1990) Pumilacidin, a complex of new antiviral antibiotics. Production, isolation, chemical properties, structure and biological activity. J Antibiot (Tokyo) 43:267–280. doi:10.7164/antibiotics.43.267
Nerurkar AS (2010) Structural and Molecular Characteristics of Lichenysin and Its Relationship with Surface Activity. In: Sen R (ed) Biosurfactants, vol 672. New York, Springer, pp 304–315. doi:10.1007/978-1-4419-5979-9_23
Ongena M, Jacques P (2008) Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol 16:115–125. doi:10.1016/j.tim.2007.12.009
Pal RR, Khardenavis AA, Purohit HJ (2015) Identification and monitoring of nitrification and denitrification genes in Klebsiella pneumoniae EGD-HP19-C for its ability to perform heterotrophic nitrification and aerobic denitrification. Funct Integr Genomics 15:63–76. doi:10.1007/s10142-014-0406-z
Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL (2007) Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6:29–40. doi:10.1038/nrd2201
Prakash O, Green SJ, Jasrotia P, Ovreholt WA, Canion A, Watson DB, Brooks SC, Kostka JE (2012) Rhodanobacter denitrificans sp. nov., isolated from nitrate-rich zones of a contaminated aquifer. Int J Syst Evol Microbiol 62:2457–2462. doi:10.1099/ijs.0.035840-0
Raaijmakers JM, de Bruijn I, Nybroe O, Ongena M (2010) Natural functions of lipopeptides from bacillus and pseudomonas: More than surfactants and antibiotics. FEMS Microbiol Rev 34:1037–1062. doi:10.1111/j.1574-6976.2010.00221.x
Rey MW, Ramaiya P, Nelson BA, Brody-Karpin SD, Zaretsky EJ, Tang M, Lopez de Leon A, Xiang H, Gusti V, Clausen IG et al (2004) Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related Bacillus species. Genome Biol 5:R77. doi:10.1186/gb-2004-5-10-r77
Richter M, Rosselló-Móra R, Glöckner FO, Peplies J (2015) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–31. doi:10.1093/bioinformatics/btv681
Rodrigues L, Banat IM, Teixeira J, Oliveira R (2006) Biosurfactants: potential applications in medicine. J Antimicrob Chemother 57:609–618. doi:10.1093/jac/dkl024
Satpute SK, Banpurkar AG, Dhakephalkar PK, Banat IM, Chopade BA (2010) Methods for investigating biosurfactants and bioemulsifiers: a review. Crit Rev Biotechnol 30:127–144. doi:10.3109/07388550903427280
Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M (2006) ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–6. doi:10.1093/nar/gkj014
Van Lanen SG, Shen B (2006) Microbial genomics for the improvement of natural product discovery. Curr Opin Microbiol 9:252–260. doi:10.1016/j.mib.2006.04.002
Villanueva J, Switala J, Ivancich A, Loewen PC (2014) Genome sequence of Bacillus pumilus MTCC B6033. Genome Announc 2:e00327–14. doi:10.1128/genomeA.00327-14
Wang H, Fewer DP, Holm L, Rouhiainen L, Sivonen K (2014) Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci U S A 111:9259–9264. doi:10.1073/pnas.1401734111
Weber T, Blin K, Duddela S, Krug D, Hu K, Bruccoleri R, Lee SY, Fischback MA, Muller R, Wohlleben W, Breitling R, Takano E, Medema MH (2015) antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:1–7. doi:10.1093/nar/gkv437
Yakimov MM, Timmis KN, Wray V, Fredrickson HL (1995) Characterization of a new lipopeptide surfactant produced by thermotolerant and halotolerant subsurface Bacillus licheniformis BAS50. Appl Environ Microbiol 61:1706–1713, doi: 0099-2240/95/$04.0010
Zerikly M, Challis GL (2009) Strategies for the discovery of new natural products by genome mining. ChemBioChem 10:625–633. doi:10.1002/cbic.200800389
Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS (2011) PHAST: a fast phage search tool. Nucleic Acids Res 39:1–6. doi:10.1093/nar/gkr485
Ziemert N, Podell S, Penn K, Badger JH, Allen E, Jensen PR (2012) The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One 7:1–9. doi:10.1371/journal.pone.0034064
Acknowledgments
Authors are thankful to Dr. Kiran Mahale (National Centre for Cell Science, Pune) and Dr. Sneha Sagarkar (Department of Biotechnology, Savitribai Phule University of Pune) for their valuable technical inputs. Authors are grateful to Mrs. Ashwini Rane (Department of Microbiology, Savitribai Phule University of Pune) for her assistance in conducting biosurfactant assay, and Datar Genetics Limited, Nashik, India for their support in Genome sequencing. We would also like to thank Dr. Ranadhir Chakraborty (North Bengal University) for providing the MDR isolates used in the study. This study was funded by Department of Biotechnology, Government of India, under the project of Microbial Culture Collection (Grant number: BT/PR10054/NDB/52/94/2007).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
An erratum to this article is available at http://dx.doi.org/10.1007/s10142-016-0528-6.
Rights and permissions
About this article
Cite this article
Shaligram, S., Kumbhare, S.V., Dhotre, D.P. et al. Genomic and functional features of the biosurfactant producing Bacillus sp. AM13. Funct Integr Genomics 16, 557–566 (2016). https://doi.org/10.1007/s10142-016-0506-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-016-0506-z