Abstract
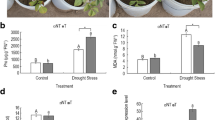
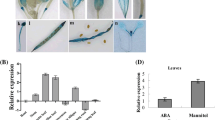
Abscisic acid (ABA)-, stress-, and ripening-induced (ASR) proteins are involved in abiotic stress responses. However, the exact molecular mechanism underlying their function remains unclear. In this study, we report that MaASR expression was induced by drought stress and MaASR overexpression in Arabidopsis strongly enhanced drought stress tolerance. Physiological analyses indicated that transgenic lines had higher plant survival rates, seed germination rates, and leaf proline content and lower water loss rates (WLR) and malondialdehyde (MDA) content. MaASR-overexpressing lines also showed smaller leaves and reduced sensitivity to ABA. Further, microarray and chromatin immunoprecipitation-based sequencing (ChIP-seq) analysis revealed that MaASR participates in regulating photosynthesis, respiration, carbohydrate and phytohormone metabolism, and signal transduction to confer plants with enhanced drought stress tolerance. Direct interactions of MaASR with promoters for the hexose transporter and Rho GTPase-activating protein (RhoGAP) genes were confirmed by electrophoresis mobility shift array (EMSA) analysis. Our results indicate that MaASR acts as a crucial regulator of photosynthesis, respiration, carbohydrate and phytohormone metabolism, and signal transduction to mediate drought stress tolerance.








Similar content being viewed by others
References
Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Sci 311:91–93
Amitai-Zeigersona H, Scolnik P, Bar-Zvi D (1995) Tomato Asrl mRNA and protein are transiently expressed following salt stress, osmotic stress and treatment with abscisic acid. Plant Sci 110:205–213
Arenhart RA, Bai Y, Valter de Oliveira LF, Bucker Neto L, Schunemann M, Maraschin Fdos S, Mariath J, Silverio A, Sachetto-Martins G, Margis R, Wang ZY, Margis-Pinheiro M (2013a) New insights into aluminum tolerance in rice: the ASR5 protein binds the STAR1 promoter and other aluminum-responsive genes. Mol Plant 7:709–721
Arenhart RA, Lima JC, Pedron M, Carvalho FE, Silveira JA, Rosa SB, Caverzan A, Andrade CM, Schünemann M, Margis R, Margis-Pinheiro M (2013b) Involvement of ASR genes in aluminium tolerance mechanisms in rice. Plant Cell Environ 36:52–67
Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24:23–58
Bartoli CG, Gomez F, Gergoff G, Guiamet JJ, Puntarulo S (2005) Up-regulation of the mitochondrial alternative oxidase pathway enhances photosynthetic electron transport under drought conditions. J Exp Bot 56:1269–1276
Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water stress studies. Plant Soil 39:205–208
Çakir B, Agasse A, Gaillard C, Saumonneau A, Delrot S, Atanassova R (2003) A grape ASR protein involved in sugar and abscisic acid signaling. Plant Cell 15:2165–2180
Cao WH, Liu J, Zhou QY, Cao YR, Zheng SF, Du BX, Zhang JS, Chen SY (2006) Expression of tobacco ethylene receptor NTHK1 alters plant responses to salt stress. Plant Cell Environ 29:1210–1219
Carmak I, Horst JH (1991) Effects of aluminum on lipid peroxidation, superoxide dismutase, catalase, and peroxidase activities in root tips of soybean (Glycine max). Physiol Plant 83:463–468
Carmo-Silva AE, Keys AJ, Andralojc PJ, Powers SJ, Arrabaça MC, Parry MA (2010) Rubisco activities, properties, and regulation in three different C4 grasses under drought. J Exp Bot 61:2355–2366
Carrari F, Fernie AR, Iusem ND (2004) Heard it through the grapevine? ABA and sugar cross-talk: the ASR story. Trends Plant Sci 9:57–59
Chaves MM (1991) Effects of water deficits on carbon assimilation. J Exp Bot 42:1–16
Chaves MM, Flexas J, Pinheiro C (2009) Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot 103:551–560
Chen JY, Liu DJ, Jiang YM, Zhao ML, Shan W, Kuang JF, Lu WJ (2011) Molecular characterization of a strawberry FaASR gene in relation to fruit ripening. PLoS One 6:e24649
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Dai JR, Liu B, Feng DR, Liu HY, He YM, Qi KB, Wang HB, Wang JF (2011) MpAsr encodes an intrinsically unstructured protein and enhances osmotic tolerance in transgenic Arabidopsis. Plant Cell Rep 30:1219–1230
Dominguez PG, Frankel N, Mazuch J, Balbo I, Iusem N, Fernie AR, Carrari F (2013) ASR1 mediates glucose-hormone cross talk by affecting sugar trafficking in tobacco plants. Plant Physiol 161:1486–1500
Ghassemian M, Nambara E, Cutler S, Kawaide Y, Kamiya Y, McCourt P (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12:1117–1126
Gonzalez-Meler MA, Matamala R, Penuelas J (1997) Effects of prolonged drought stress and nitrogen deficiency on the respiratory O2 uptake of bean and pepper leaves. Photosynthetica 34:505–512
Hall BP, Shakeel SN, Amir M, Ul Haq N, Qu X, Schaller GE (2012) Histidine kinase activity of the ethylene receptor ETR1 facilitates the ethylene response in Arabidopsis. Plant Physiol 159:682–695
Henry IM, Carpentier SC, Pampurova S, Van Hoylandt A, Panis B, Swennen R, Remy S (2011) Structure and regulation of the Asr gene family in banana. Planta 234:785–798
Hsu YF, Yu SC, Yang CY, Wang CS (2011) Lily ASR protein-conferred cold and freezing resistance in Arabidopsis. Plant Physiol Biochem 49:937–945
Hu W, Huang C, Deng X, Zhou S, Chen L, Li Y, Wang C, Ma Z, Yuan Q, Wang Y, Cai R, Liang X, Yang G, He G (2013) TaASR1, a transcription factor gene in wheat, confers drought stress tolerance in transgenic tobacco. Plant Cell Environ 36:1449–1464
Huang JC, Lin SM, Wang CS (2000) A pollen-specific and desiccation-associated transcript in Lilium longiflorum during development and stress. Plant Cell Physiol 41:477–485
Joo J, Lee YH, Kim YK, Nahm BH, Song SI (2013) Abiotic stress responsive rice ASR1 and ASR3 exhibit different tissue-dependent sugar and hormone-sensitivities. Mol Cells 35:421–435
Kalifa Y, Gilad A, Konrad Z, Zaccai M, Scolnik PA, Bar-Zvi D (2004) The water- and salt-stress-regulated Asr1 (abscisic acid stress ripening) gene encodes a zinc-dependent DNA-binding protein. Biochem J 381:373–378
Kaufmann K, Muino JM, Osteras M, Farinelli L, Krajewski P, Angenent GC (2010) Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat Protoc 5:457–472
Knoch E, Dilokpimol A, Tryfona T, Poulsen CP, Xiong G, Harholt J, Petersen BL, Ulvskov P, Hadi MZ, Kotake T, Tsumuraya Y, Pauly M, Dupree P, Geshi N (2013) A β-glucuronosyltransferase from Arabidopsis thaliana involved in biosynthesis of type II arabinogalactan has a role in cell elongation during seedling growth. Plant J 76:1016–1029
Konrad Z, Bar-Zvi D (2008) Synergism between the chaperone-like activity of the stress regulated ASR1 protein and the osmolyte glycine-betaine. Planta 227:1213–1219
Liu HY, Dai JR, Feng DR, Liu B, Wang HB, Wang JF (2010) Characterization of a novel plantain Asr gene, MpAsr, that is regulated in response to infection of Fusarium oxysporum f. sp. cubense and abiotic stresses. J Integr Plant Biol 52:315–323
Liu J, Jia C, Dong F, Wang J, Zhang J, Xu Y, Xu B, Jin Z (2013) Isolation of an abscisic acid senescence and ripening inducible gene from litchi and functional characterization under water stress. Planta 237:1025–1036
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408
Nagano AJ, Matsushima R, Hara-Nishimura I (2005) Activation of an ER-body-localized beta-glucosidase via a cytosolic binding partner in damaged tissues of Arabidopsis thaliana. Plant Cell Physiol 46:1140–1148
Ouyang B, Yang T, Li H, Zhang L, Zhang Y, Zhang J, Fei Z, Ye Z (2007) Identification of early salt stress response genes in tomato root by suppression subtractive hybridization and microarray analysis. J Exp Bot 58:507–520
Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, Bao W, Fang H, Kawasaki ES, Hager J, Tikhonova IR, Walker SJ, Zhang L, Hurban P, de Longueville F, Fuscoe JC, Tong W, Shi L, Wolfinger RD (2006) Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat Biotechnol 24:1140–1150
Ribas-Carbo M, Taylor NL, Giles L, Busquets S, Finnegan PM, Day DA, Lambers H, Medrano H, Berry JA, Flexas J (2005) Effects of water stress on respiration in soybean leaves. Plant Physiol 139:466–473
Ricardi MM, Guaimas FF, González RM, Burrieza HP, López-Fernández MP, Jares-Erijman EA, Estévez JM, Iusem ND (2012) Nuclear import and dimerization of tomato ASR1, a water stress-inducible protein exclusive to plants. PLoS One 7:e41008
Ricardi MM, González RM, Zhong S, Domínguez PG, Duffy T, Turjanski PG, Salgado Salter JD, Alleva K, Carrari F, Giovannoni JJ, Estévez JM, Iusem ND (2014) Genome-wide data (ChIP-seq) enabled identification of cell wall-related and aquaporin genes as targets of tomato ASR1, a drought stress-responsive transcription factor. BMC Plant Biol. doi:10.1186/1471-2229-14-29
Saumonneau A, Laloi M, Lallemand M, Rabot A, Atanassova R (2012) Dissection of the transcriptional regulation of grape ASR and response to glucose and abscisic acid. J Exp Bot 63:1495–1510
Shkolnik D, Bar-Zvi D (2008) Tomato ASR1 abrogates the response to abscisic acid and glucose in Arabidopsis by competing with ABI4 for DNA binding. Plant Biotechnol J 6:368–378
Sreedharan S, Shekhawat UK, Ganapathi TR (2013) Transgenic banana plants overexpressing a native plasma membrane aquaporin MusaPIP1;2 display high tolerance levels to different abiotic stresses. Plant Biotechnol J 11:942–952
Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29:417–426
Virlouvet L, Jacquemot MP, Gerentes D, Corti H, Bouton S, Gilard F, Valot B, Trouverie J, Tcherkez G, Falque M, Damerval C, Rogowsky P, Perez P, Noctor G, Zivy M, Coursol S (2011) The ZmASR1 protein influences branched-chain amino acid biosynthesis and maintains kernel yield in maize under water-limited conditions. Plant Physiol 157:917–936
Voxeur A, André A, Breton C, Lerouge P (2012) Identification of putative rhamnogalacturonan-II specific glycosyltransferases in Arabidopsis using a combination of bioinformatics approaches. PLoS One 7:e51129
Xu BY, Su W, Liu JH, Wang JB, Jin ZQ (2007) Differentially expressed cDNAs at the early stage of banana ripening identified by suppression subtractive hybridization and cDNA microarray. Planta 226:529–539
Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30:e15
Yang CY, Chen YC, Jauh GY, Wang CS (2005) A lily ASR protein involves abscisic acid signaling and confers drought and salt resistance in Arabidopsis. Plant Physiol 139:836–846
Yu XM, Griffith M, Wiseman SB (2001) Ethylene induces antifreeze activity in winter rye leaves. Plant Physiol 126:1232–1240
Zhao XC, Schaller GE (2004) Effect of salt and osmotic stress upon expression of the ethylene receptor ETR1 in Arabidopsis thaliana. FEBS Lett 562:189–192
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31071788), the earmarked fund for Modern Agro-industry Technology Research System (CARS-32), the Ministry of Science and Technology of the People’s Republic of China (2011AA10020605), and the Major Technology Project of Hainan (ZDZX2013023-1).
Authors’ contributions
ZQJ and BYX conceived the study. LLZ, YW, RJF, YDZ, JHL, CHJ, HXM, and JBZ performed the experiments and carried out the analysis. WH designed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Lili Zhang and Wei Hu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
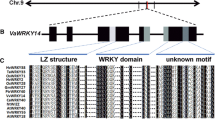
Alignment of MaASR sequence with homologues from other species: mASR1 (GU134740) from Musa acuminata subsp, mASR2 (GU134761) from Musa balbisiana, mASR4 (GU134735) from Musa banksii, TaASR1 (HQ287799) from Triticum aestivum, ZmASR2 (NP_001147703.1) from Zea mays, OsASR1 (AAX92999.1) from Oryza sativa. ABA/WDS motif was displayed in the box. Letters marked with double transverse lines showed a putative nuclear targeting signal. One His-rich region and two Ala-rich regions are underlined. Amino acid sequences alignment was performed by ClusterX 2.0. (GIF 79 kb)
Fig. S2
Phylogenetic analysis of MaASR with RcASR (XP_002524296.1) from Ricinus communis, VpASR (ABC86744.1) from Vitis pseudoreticulata, SlASR4 (AAY98026.1) from Solanum lycopersicum, ScASR4 (AAY98030.1) from Solanum corneliomuelleri, PpASR (BAA96451.1) from Pyrus pyrifolia, SpASR2 (ABY84856.1) from Solanum pimpinellifolium, SlASR2 (ABY84857.1) from Solanum lycopersicum, LpASR2 (ABS19518.1) from Lycopersicon peruvianum, NbASR (ACV52581.1) from Nicotiana benthamiana, mASR1 (GU134740) from Musa acuminata subsp. Burmannicoides, mASR2 (GU134761) from Musa balbisiana, mASR4 (GU134735) from Musa banksii, ZmASR2 (NP_001147703.1) from Zea mays, OsASR (AAX92999.1) from Oryza sativa, TaASR1 (HQ287799) from Triticum aestivum, LlASR (AAM51877.1) from Lilium longiflorum. Phylogenetic tree was constructed by using ClusterX 2.0 and Mega 4.0. (GIF 15 kb)
Fig. S3
Cluster data (A) and scatter plot (B) of differentially expressed genes identified by microarrays. (A) The red color represents upregulated genes with fold change ≥ 2, with screening criteria ratio ≥ 2; The green color represents downregulated genes with fold change ≥ 2 and screening criteria ratio ≤ 0.5. (B) X and Y axes represent the fluorescence signal intensity value of the WT and transgenic lines, respectively. Each point represents the hybridization signal of the gene in the expression profile microarray. The red color represents the genes with a screening criteria ratio ≥ 2, while the green color represents genes with a screening criteria ratio ≤ 0.5; The black color represents genes that have no significant changes with screening criteria of 0.5 < ratio < 2. (GIF 77 kb)
Fig. S4
Expression of phytohormone-metabolic/-transduction-related genes determined by qRT-PCR analysis in WT and transgenic lines. Fourteen day old Arabidopsis seedlings of each sample were transferred to filter paper for 2 h or 6 h to induce dehydration. The fold difference in mRNA was relative to that of WT under normal conditions. Vertical bars indicate ± SE of three independent measurements. (GIF 95 kb)
Fig. S5
Categorization of the differentially expressed genes identified from the Arabidopsis expression profile microarray (14 vs WT) according to position in the cellular component (A), molecular biological function (B) and biological processes (C). (GIF 82 kb)
Fig. S6
Categorization of the differentially expressed genes identified from the Arabidopsis expression profile microarray (Y vs W) according to position in the cellular component (A), molecular biological function (B) and biological processes (C). (GIF 90 kb)
Fig. S7
Analysis of the specificity of the MaASR monoclonal antibody used for chromatin immunoprecipitation (ChIP) assay. (A) SDS-PAGE analysis of expressed MaASR protein after Ni-agarose affinity chromatography. M: Protein Marker; 1: Purified protein of MaASR. (B) Western bolt analysis of the specificity of the MaASR monoclonal antibody. M: Protein marker; 1: Hybridization of MaASR monoclonal antibody with proteins expressed by vacant vector; 2: Hybridization of MaASR monoclonal antibody with purified MaASR protein. (GIF 22 kb)
Fig. S8
The expression of hexose transporter and RhoGAP genes determined by qRT-PCR analysis in WT and transgenic lines. Fourteen day old Arabidopsis seedlings of each sample were transferred to filter paper for 2 h or 6 h to induce dehydration. The fold difference in mRNA was relative to that of WT under normal conditions. Vertical bars indicate ± SE of three independent measurements. (GIF 25 kb)
Fig. S9
Network diagram (A) and connectivity (B) of differentially expressed genes in Y vs W. (A) Red represents upregulated genes and yellow represents downregulated genes. CX = mRNA co-expression between Arabidopsis genes; DC = Domain co-occurrence between Arabidopsis proteins; GN = Gene neighborhoods between Arabidopsis orthologs in bacterial genomes; LC = Literature-curated Arabidopsis protein interactions; PG = Phylogenetic profile similarity between Arabidopsis homologues. (B) The horizontal axis represents genes ID in the network diagram. The vertical axis represents gene connectivity in network diagram. (GIF 96 kb)
Table S1
qRT-PCR primers used in this study. (XLS 26 kb)
Table S2
Upregulated genes in the expression profile microarray (14 vs WT). (XLS 114 kb)
Table S3
Downregulated genes in the expression profile microarray (14 vs WT). (XLS 54 kb)
Table S4
Upregulated genes in the expression profile microarray (Y vs W). (XLS 72 kb)
Table S5
Downregulated genes in the expression profile microarray (Y vs W). (XLS 89 kb)
Table S6
Differentially expressed genes related to water deprivation, photosynthesis, respiration, carbohydrate metabolism, amino acid metabolism and phytohormone metabolism and signal transduction in the expression profile microarray (14 vs WT). (XLS 70 kb)
Table S7
Differentially expressed genes related to water deprivation, photosynthesis, respiration, carbohydrate metabolism, amino acid metabolism and phytohormone metabolism and signal transduction in the expression profile microarray (Y vs W). (XLS 79 kb)
Table S8
Summary of genes and binding sites directly targeted by MaASR. (XLS 190 kb)
Table S9
Genes directly targeted by MaASR identified by comparing ChIP-seq and microarray (Y vs W) data. (XLS 24 kb)
Table S10
Statistics of sugar-responsive elements SURE1 (AATAGAAAA), sucrose box 3 (AAAATCA---TAA) and CE1 (CACCG) among genes directly targeted by MaASR. (XLS 28 kb)
Table S11
Corresponding genes according to GO enrichment analysis of MaASR directly regulated genes. (XLS 32 kb)
Table S12
(DOC 41 kb)
Rights and permissions
About this article
Cite this article
Zhang, L., Hu, W., Wang, Y. et al. The MaASR gene as a crucial component in multiple drought stress response pathways in Arabidopsis . Funct Integr Genomics 15, 247–260 (2015). https://doi.org/10.1007/s10142-014-0415-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-014-0415-y