Abstract
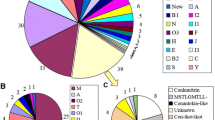
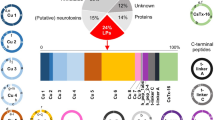
The venom of each species of Conus contains different kinds of pharmacologically active peptides which are mostly unique to that species. Collectively, the ~500–700 species of Conus produce a large number of these peptides, perhaps exceeding 140,000 different types in total. To date, however, only a small fraction of this diversity has been characterized via transcriptome sequencing. In addition, the sampling of this chemical diversity has not been uniform across the different lineages in the genus. In this study, we used high-throughput transcriptome sequencing approach to further investigate the diversity of Conus venom peptides. We chose a species, Conus tribblei, as a representative of a poorly studied clade of Conus. Using the Roche 454 and Illumina platforms, we discovered 136 unique and novel putative conopeptides belonging to 30 known gene superfamilies and 6 new conopeptide groups, the greatest diversity so far observed from a transcriptome. Most of the identified peptides exhibited divergence from the known conopeptides, and some contained cysteine frameworks observed for the first time in cone snails. In addition, several enzymes involved in posttranslational modification of conopeptides and also some proteins involved in efficient delivery of the conopeptides to prey were identified as well. Interestingly, a number of conopeptides highly similar to the conopeptides identified in a phylogenetically distant species, the generalist feeder Conus californicus, were observed. The high diversity of conopeptides and the presence of conopeptides similar to those in C. californicus suggest that C. tribblei may have a broad range of prey preferences.







Similar content being viewed by others
References
Aguilar MB, Zugasti-Cruza A, Falcóna A, Batista CVF, Olivera BM, Heimer de la Coter EP (2013) A novel arrangement of Cys residues in a paralytic peptide of Conus cancellatus (jr. syn.: Conus austini), a worm-hunting snail from the Gulf of Mexico. Peptides 41:38–44
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Apweiler R, Bairoch A, Wu CH (2004) Protein sequence databases. Curr Opin Chem Biol 8:76–80
Bayrhuber M, Vijayan V, Ferber M, Graf R, Korukottu J, Imperial J, Garrett JE, Olivera BM, Terlau H, Zweckstetter M, Becker S (2005) Conkunitzin-S1 is the first member of a new kunitz-type neurotoxin family, structural and functional characterization. J Biol Chem 280:23766–23770
Biggs JS, Olivera BM, Kantor YI (2008) α-conopeptides specifically expressed in the salivary gland of Conus pulicarius. Toxicon 52:101–105
Biggs JS, Watkins M, Puillandre N, Ownby J, Christensen S, Moreno, E L-V, Christensen S, Moreno KJ, Navarro AL, Corneli PS, Olivera BM (2010) Evolution of conus peptide toxins: analysis of Conus californicus Reeve, 1844. Mol Phylogenet Evol 56:1–12
Campanella JJ, Bitincka L, Smalley J (2003) MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinforma 4:29
Chen P, Garrett JE, Watkins M, Olivera BM (2008) Purification and characterization of a novel excitatory peptide from Conus distans venom that defines a novel gene superfamily of conotoxins. Toxicon 52:139–145
Conticello SG, Gilad Y, Avidan N, Ben-Asher E, Levy Z, Fainzilber M (2001) Mechanisms for evolving hypervariability: the case of conopeptides. Mol Biol Evol 18:120–131
Duckert P, Brunak S, Blom N (2004) Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel 17:107–112
Duda TF, Palumbi SR (2004) Gene expression and feeding ecology: evolution of piscivory in the venomous gastropod genus Conus. Proc R Soc London B 271:1165–1174
Duda TF, Chang D, Lewis BD, Lee T (2009) Geographic variation in venom allelic composition and diets of the widespread predatory marine gastropod Conus ebraeus. PLoS One 4:e6245
Dutertre S, Biass D, Stöcklin R, Favreau P (2010) Dramatic intraspecimen variations within the injected venom of Conus consors: an unsuspected contribution to venom diversity. Toxicon 55:1453–1462
Dutertre S, Jin A, Kaas Q, Jones A, Alewood PF, Lewis RJ (2013) Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol Cell Proteomics 12:312–329
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Elliger CA, Richmond TA, Lenaric ZN, Pierce NT, Sweedler JV, Gilly WF (2011) Diversity of conotoxin types from Conus californicus reflects a diversity of prey types and a novel evolutionary history. Toxicon 57:311–322
England LJ, Imperial J, Jacobsen R, Craig AG, Gulyas J, Akhtar M, Rivier J, Julius D, Olivera BM (1998) Inactivation of a serotonin-gated ion channel by a polypeptide toxin from marine snails. Science 281:575–578
Espiritu DJ, Watkins M, Dia-Monje V, Cartier GE, Cruz LJ, Olivera BM (2001) Venomous cone snails: molecular phylogeny and the generation of toxin diversity. Toxicon 39:1899–1916
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol 29:644–654
Hall TA (1999) BioEdit, a user-friendly biological sequence alignment editor and analysis program for Windows 95, 98, NT. Nucleic Acids Symp Ser 41:95–98
Hu H, Bandyopadhyay PK, Olivera BM, Yandell M (2011) Characterization of the Conus bullatus genome and its venom-duct transcriptome. BMC Genomics 12:60
Hu H, Bandyopadhyay PK, Olivera BM, Yandell M (2012) Elucidation of the molecular envenomation strategy of the cone snail Conus geographus through transcriptome sequencing of its venom duct. BMC Genomics 13:284
Huelsenbeck JP, Ronquist F, Hall B (2001) MrBayes: bayesian inference of phylogeny. Bioinformatics 17:754–755
Jacob RB, McDougal OM (2010) The M-superfamily of conotoxins: a review. Cell Mol Life Sci 67:17–27
Kaas Q, Westermann J, Craik DJ (2010) Conopeptide characterization and classifications: an analysis using conoserver. Toxicon 55:1491–1509
Kaas Q, Yu R, Jin A, Dutertre S, Craik DJ (2012) ConoServer: updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res 40:D325–D330
Kordiŝ D, Gubenŝek F (2000) Adaptive evolution of animal toxin multigene families. Gene 261:43–52
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–360
Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, Mcwilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lavergne V, Dutertre S, Jin A, Lewis RJ, Taft RJ, Alewood PF (2013) Systematic interrogation of the Conus marmoreus venom duct transcriptome with conosorter reveals 158 novel conotoxins and 13 new gene superfamilies. BMC Genomics 14:708
Lirazan MB, Jimenez EC, Craig AG, Olivera BM, Cruz LJ (2002) Conophysin-R, a Conus radiatus venom peptide belonging to the neurophysin family. Toxicon 40:901–908
Lluisma AO, Milash BA, Moore M, Olivera BM, Bandyopadhyay PK (2012) Novel venom peptides from the cone snail Conus pulicarius discovered through next-generation sequencing of its venom duct transcriptome. Mar Genom 5:43–51
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu S, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam T, Wang J (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaSci 1(18):1–6
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer MLI, Jarvie TP, Jirage KB, Kim J, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–381
Maricq AV, Jensen S, Walker C, Madsen D, Olivera BM, Ellison M (2009) Conus polypeptides. United States Patent US2010/0197567 A1
McIntosh JM, Ghomashchi F, Gelb MH, Dooley DJ, Stoehr SJ, Giordani AB, Naisbitt SR, Olivera BM (1995) Conodipine-M, a novel phospholipase A2 isolated from the venom of the marine snail Conus magus. J Biol Chem 270:3518–3526
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–152
Olivera BM (2006) Conus peptides: biodiversity-based discovery and exogenomics. J Biol Chem 281:31173–31177
Olivera BM, McIntosh JM, Cruz LJ, Luque FA, Gray WR (1984) Purification and sequence of a presynaptic peptide toxin from Conus geographus venom. Biochemistry 22:5078–5090
Olivera BM, Teichert RW (2007) Diversity of the neurotoxin Conus peptides, a model for concerted pharmacological discovery. Mol Interven 7:251–260
Olivera BM, Corneli PS, Watkins M, Fedosov A (2014) Biodiversity of cone snails and other venomous marine gastropods: evolutionary success through neuropharmacology. Annu Rev Anim Biosci 2:487–513
Pearson WR, Wood T, Zhang Z, Miller W (1997) Comparison of DNA sequences with protein sequences. Genomics 46:24–36
Peng C, Liu L, Shao X, Chi C, Wang C (2008) Identification of a novel class of conotoxins defined as V-conotoxins with a unique cysteine pattern and signal peptide sequence. Peptides 29:985–991
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Puillandre N, Koua D, Favreau P, Olivera BM, Stöcklin R (2012) Molecular phylogeny, classification and evolution of conopeptides. J Mol Evol 74:297–309
Rambaut A, Drummond AJ (2007) Tracer v1.4, Available from http://beast.bio.ed.ac.uk/Tracer
Rawlings ND, Tolle DP, Barrett AJ (2004) Evolutionary families of peptidase inhibitors. Biochem J, 378: 705–716
Remigio EA, Duda TF (2008) Evolution of ecological specialization and venom of a predatory marine gastropod. Mol Ecol 17:1156–1162
Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, Griffith M, Raymond A, Thiessen N, Cezard T, Butterfield YS, Newsome R, Chan SK, She R, Varhol R, Kamoh B, Prabhu A, Tam A, Zhao Y, Moore RA, Hirst M, Marra MA, Jones SJM, Hoodless PA, Birol I (2010) De novo assembly and analysis of RNA-seq data. Nat Methods 7:909–915
Safavi-Hemami H, Siero WA, Kuang Z, Williamson NA, Karas JA, Page LR, MacMillan D, Callaghan B, Kompella SN, Adams DJ, Norton RS, Purcell AW (2011) Embryonic toxin expression in the cone snail Conus victoriae: primed to kill or divergent function? J Biol Chem 286:22546–22557
Safavi-hemami H, Gorasia DG, Steiner AM, Williamson NA, Karas JA, Gajewiak J, Olivera BM, Bulaj G, Purcell AW (2012) Modulation of conotoxin structure and function is achieved through a multienzyme complex in the venom ducts of cone snails. J Biol Chem 287:34288–34303
Schulz MH, Zerbino DR, Vingron M, Birney E (2012) Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28:1086–1092
Smit AB, van Marle A, van Elk R, Bogerd J, van Heerikhuizen H, Geraerts WP (1993) Evolutionary conservation of the insulin gene structure in invertebrates: cloning of the gene encoding molluscan insulin-related peptide III from Lymnaea stagnalis. J Mol Endocrinol 11:103–113
Terrat Y, Biass D, Dutertre S, Favreau P, Remm M, Stöcklin R, Piquemal D, Ducancel F (2012) High-resolution picture of a venom duct transcriptome: case study with the marine snail Conus consors. Toxicon 59:34–46
Van Kesteren RE, Smit AB, De Lange RP, Kits KS, Van Golen FA, Van Der Schors RC, De With ND, Burke JF, Geraerts WP (1995) Structural and functional evolution of the vasopressin/oxytocin superfamily: vasopressin-related conopressin is the only member present in Lymnaea, and is involved in the control of sexual behavior. J Neurosci 15:5989–5998
Walker CS, Jensen S, Ellison M, Matta JA, Lee WY, Imperial SJ, Duclos N, Brockie PJ, Madsen DM, Isaac JTR, Olivera BM, Maricq AV (2010) A novel Conus snail polypeptide causes excitotoxicity by blocking desensitization of AMPA receptors. Curr Biol 19:900–908
Wang Z, Han Y, Shao X, Chi C, Guo Z (2007) Molecular cloning, expression and characterization of protein disulfide isomerase from Conus marmoreus. FEBS J 274:4778–4787
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Boloud L, Wang J (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34(Web service issue):293–297
Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H, Xiong Z, Que H, Xie Y, Holland PWH, Paps J, Zhu Y, Wu F, Chen Y, Wang J, Peng C, Meng J, Yang L, Liu J, Wen B, Zhang N, Huang Z, Zhu Q, Feng Y, Mount A, Hedgecock D, Xu Z, Liu Y, Domazet-Lošo T, Du Y, Sun X, Zhang S, Liu B, Cheng P, Jiang X, Li J, Fan D, Wang W, Fu W, Wang T, Wang B, Zhang J, Peng Z, Li Y, Li N, Wang J, Chen M, He Y, Tan F, Song X, Zheng Q, Huang R, Yang H, Du X, Chen L, Yang M, Gaffney PM, Wang S, Luo L, She Z, Ming Y, Huang W, Zhang S, Huang B, Zhang Y, Qu T, Ni P, Miao G, Wang J, Wang Q, Steinberg CEW, Wang H, Li N, Qian L, Zhang G, Li Y, Yang H, Liu X, Wang J, Yin Y, Wang J (2012) The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490:49–54
Acknowledgments
The specimens used in this study were obtained in conjunction with a collection trip supported in part by ICBG grant 1U01TW008163. The Illumina sequencing performed in Oregon Health and Science University was funded by the Philippine PharmaSeas Drug Discovery Program. Roche 454 sequencing was supported by NIH grant GM48677 (BMO) and performed in Marine Science Institute, University of the Philippines. This study was partially supported by the Office of the Vice President for Academic Affairs, University of the Philippines through Philippine Genome Center. The data analysis was carried out using the High-Performance Computing Facility of the Advanced Science and Technology Institute and the Philippine e-Science Grid, Diliman, Quezon City. We would like to thank Joeriggo Reyes for the help in transcriptome analysis and Alexander Fedosov for the help in phylogenetic analysis. We would like to thank Maren Watkins for reviewing the conopeptide sequences and for her constructive comments.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 27 kb)
Fig. S 1
The putative conopeptide precursors identified by ConoSorter. The prepeptide cleavage sites are shown in bold and are underlined (DOC 31 kb)
Fig. S 2
The truncated putative conopeptide precursors of (a) A-, (b) G- and (c) O1-superfamilies. The conopeptides identified in C. tribblei are shown in black and the conopeptide nomenclature is as described in "Materials and Methods". The name of each conopeptide is presented as Ctr_#_T/N/TN: Ctr: C. tribblei, #: arbitrary assigned number, T: only present in the ‘Trinity conopeptide dataset’, N: only present in the ‘Newbler conopeptide dataset’, TN: present in both Trinity and Newbler conopeptide datasets. The reference sequences are shown in green and cysteine residues are shown in bold italic red. The names of the reference sequences are derived from the ConoServer database unless noted otherwise. The mature regions are underlined and the signal regions are highlighted (DOC 35 kb)
Fig. S 3
The putative conopeptide precursors of (a) M superfamily, (b) con-ikot-ikot and (c) conkunitzin family. The notes are indicated in Fig. S 2 (DOC 46 kb)
Fig. S 4
The putative conopeptide precursors of (a) conopressin/conophysin and (b) conodipine families. Conopressin domain is underlined in red at position 32–40 and conophysin is underlined in gray. The notes are indicated in Fig. S 2 (DOC 45 kb)
Fig. S 5
The putative conopeptide precursors of (a) N-, (b) K- and (c) S-superfamilies. The notes are indicated in Fig. S 2 (DOC 36 kb)
Fig. S 6
The putative conopeptide precursors of (a) I2-, (b) L-, (c) H-, (d) O1- and (e) O2-superfamilies. The notes are indicated in Fig. S 2 (DOC 50 kb)
Fig. S 7
The putative conopeptide precursors of (a) D-, (b) I1- and (c) I3-superfamilies. The notes are indicated in Fig. S 2 (DOC 34 kb)
Rights and permissions
About this article
Cite this article
Barghi, N., Concepcion, G.P., Olivera, B.M. et al. High Conopeptide Diversity in Conus tribblei Revealed Through Analysis of Venom Duct Transcriptome Using Two High-Throughput Sequencing Platforms. Mar Biotechnol 17, 81–98 (2015). https://doi.org/10.1007/s10126-014-9595-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-014-9595-7