Abstract
Acute undifferentiated fever (AUF) poses a diagnostic challenge due to the variety of possible aetiologies. While the majority of AUFs resolve spontaneously, some cases become prolonged and cause significant morbidity and mortality, necessitating improved diagnostic methods. This study evaluated the utility of deep sequencing in fever investigation. DNA and RNA were isolated from plasma/sera of AUF cases being investigated at Cairns Hospital in northern Australia, including eight control samples from patients with a confirmed diagnosis. Following isolation, DNA and RNA were bulk amplified and RNA was reverse transcribed to cDNA. The resulting DNA and cDNA amplicons were subjected to deep sequencing on an Illumina HiSeq 2000 platform. Bioinformatics analysis was performed using the program Kraken and the CLC assembly-alignment pipeline. The results were compared with the outcomes of clinical tests. We generated between 4 and 20 million reads per sample. The results of Kraken and CLC analyses concurred with diagnoses obtained by other means in 87.5 % (7/8) and 25 % (2/8) of control samples, respectively. Some plausible causes of fever were identified in ten patients who remained undiagnosed following routine hospital investigations, including Escherichia coli bacteraemia and scrub typhus that eluded conventional tests. Achromobacter xylosoxidans, Alteromonas macleodii and Enterobacteria phage were prevalent in all samples. A deep sequencing approach of patient plasma/serum samples led to the identification of aetiological agents putatively implicated in AUFs and enabled the study of microbial diversity in human blood. The application of this approach in hospital practice is currently limited by sequencing input requirements and complicated data analysis.
Similar content being viewed by others
Introduction
Acute undifferentiated fever (AUF) is caused by a variety of causes, producing a range of clinical manifestations with acute fever as a unifying symptom. Most clinicians and researchers define acute fever as evidence of raised body temperature to ≥38 °C for ≤3 weeks, without detection of systemic disease or the focus of infection or inflammation after initial clinical evaluation and basic laboratory investigations [1]. This condition poses a diagnostic challenge for clinicians due to non-specific clinical features and the indistinctive profile of routine blood tests. Without a specific aetiological diagnosis, the treatment of AUF is often based on an ‘educated guess’ or a syndromic approach that often leads to inappropriate treatment [2, 3].
It has long been known that infection is the main cause of fever, particularly in the acute stage. Unfortunately, there are hundreds of possible aetiologies of fever, such that conventional diagnostic tools are often either unavailable or restricted to a subset of the ‘most likely’ infectious agents due to the high costs associated with laboratory testing. The limitation of the current diagnostic approach causes a significant proportion of fever to go undiagnosed. Indeed, the frequencies of undiagnosed AUFs in Asian tropical countries ranges from 8 to 80 % [1].
The wide availability of nucleic acid [i.e. polymerase chain reaction (PCR)-based] assays in clinical laboratories provides sensitive and specific detection of pathogens. However, while techniques such as multiplex PCR can provide simultaneous detection of multiple pathogens, this approach is impractical for more than a handful of pathogens in any one assay [4, 5] and is not capable of detecting novel pathogens [6]. Diagnostic microarrays can expand detection capacity considerably, allowing simultaneous detection of tens of pathogens or more [7, 8], but these too are limited for the detection of novel or emerging pathogens [9].
The advent of next-generation sequencing (NGS) [10] provides a basis for unbiased identification of infectious agents associated with AUF, as well as the capacity to identify novel and emerging pathogens [9, 11, 12]. This study aimed to evaluate the practical use of NGS technology as a diagnostic tool for the identification of infectious agents causing fever. This method is referred to as metagenomic deep sequencing and has been used in previous studies to determine the agents responsible for dengue-like illnesses [9] and acute haemorrhagic fever [13].
Materials and methods
Sample collection
We collected 40 plasma/serum samples from patients who presented to Cairns Hospital, a tertiary hospital in Cairns, Far North Queensland, Australia (16.9256° S, 145.7753° E). The inclusion criteria included patients aged 16 to 65 years who had raised temperature of ≥38 °C or history of fever with feeling cold or shivering for up to 21 days with no evident focus of infection and no obvious cause of fever after initial clinical, radiology and laboratory evaluation; this included tests for which results were normally reported within 6 h from admission. Thus, a specific diagnosis at the time of patient recruitment was unavailable. Subsequent investigation(s) determined by the attending doctors ascertained an aetiological diagnosis in a subset of study participants (control subjects), while other participants (test subjects) remained undiagnosed (Fig. 1).
Flow chart of patient selection. Definitions: Acute fever is an increase of body temperature to 38 °C or more for a period of 21 days or less. Initial investigations refer to comprehensive clinical assessment and basic laboratory and radiology tests; this included tests that are normally reported within 6 h from admission. Comprehensive clinical assessment includes complete history taking and thorough physical examination. Basic laboratory tests usually include complete blood count and urinalysis. Basic radiology tests could include chest X-ray, abdomen and pelvic X-ray, and ultrasonography. Fever with obvious likely diagnosis is any case of fever with definitive diagnosis immediately after initial investigations. This includes fever cases with an obvious focus of infection or local inflammation, such as community-acquired pneumonia, urinary tract infection, skin and soft tissue infection, bone and dental infection, pelvic inflammatory disease and intra-abdominal infection. Acute undifferentiated fever is any case of acute fever with unclear aetiology and the results of initial investigations are not conclusive in achieving a diagnosis. Thus, the condition is characterised by a requirement for further investigation to explain the cause of fever and to consider differential diagnoses. Rational diagnostic investigations are further tests as judged by an attending doctor to determine the cause of fever, such as further serology, cerebrospinal fluid analysis and/or advanced radiology tests [computed tomography (CT) scan, magnetic resonance imaging (MRI)]. Samples were collected from both groups of participants (diagnosed subjects and undiagnosed subjects) for fever investigation using the deep sequencing approach
Sample preparation and sequencing
DNA was isolated from 200 μl of plasma/serum using the QIAamp® DNA Mini Kit (Qiagen), while RNA was isolated from 250 μl of sample mixed with 750 μl of TRIzol® LS reagent (Life Technologies), as per the manufacturer’s protocol. Each sample preparation was performed in duplicate. Following RNA isolation, genomic DNA was removed from RNA samples using DNase I, Amplification Grade (Sigma Aldrich). Amplification of DNA and RNA was conducted according to the SeqPlex Enhanced DNA Amplification Kit and the SeqPlex RNA Amplification Kit (Sigma Aldrich) protocols, resulting in double-stranded DNA and cDNA amplicons, respectively. The GenElute PCR Clean-Up Kit (Sigma-Aldrich) was used for the purification of products from the SeqPlex DNA and RNA amplification kits. The quantity, size and purity of DNA/cDNA amplicons were determined using gel electrophoresis (1.5 % agarose) and a NanoDrop 2000 spectrophotometer (Thermo Scientific), as requested by the commercial sequencing service used for the study: the Australian Genome Research Facility (AGRF). The samples that passed AGRF quality assessment were processed into library preparations using the TruSeq Nano DNA Library Preparation Kit protocol (Illumina). Paired-end (PE) 100-bp sequencing was conducted using an Illumina HiSeq 2000 instrument.
Bioinformatics analysis
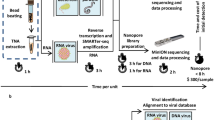
Identification of pathogens associated with AUF was performed on two cloud computing servers: BaseSpace® (Illumina) and CLC Genomics Workbench (Qiagen) (Fig. 2). First, the raw sequence data obtained from AGRF were uploaded onto the BaseSpace® server; then, human sequences in each dataset were identified by alignment to the human reference genome (hg19) using the program SNAP version 1.0beta.14 [14] and removed. The remaining sequences were then classified using the default parameters of the program Kraken version 0.10.4-beta [15], based on their homology with organisms in the MiniKraken 20140330 database, which contains a collection of complete bacterial, archaeal and viral genomes available from the National Center for Biotechnology Information (NCBI) RefSeq database.
Analysis workflow. Primary and secondary analyses were performed sequentially for all samples and performed in parallel for the purpose of validation. Sequential analysis means that all reads were uploaded to the BaseSpace® server, followed by analysis of non-human reads not classified by Kraken in the CLC server. Validation of next-generation sequencing (NGS) analysis was performed on positive control samples only by analysing all reads using the BaseSpace® and CLC servers in parallel
Any non-human reads not classified by Kraken were imported to the CLC Genomics Workbench server. The quality of the sequence data was examined using the FastQC tool [16]. The reads were then processed using the CLC Trim Sequences tool to remove adapters, low quality bases, ambiguous nucleotides, terminal nucleotides (25–35 nucleotides from the 5’ end) and short sequences (less than 24 nucleotides). Then, reads were mapped and filtered a second time using the CLC read mapper program to the human reference genome (hg19). All remaining reads were assembled using the CLC De Novo Assembly tool. Assembled contigs were compared with the NCBI non-redundant database using the Basic Local Alignment Search Tool (BLAST) [17]. The BLASTn program optimised for highly similar sequences (megablast) was used to search nucleotide databases for sequence(s) that matched a nucleotide query. The nearest matching sequence (e-value threshold ≤10−5) was accepted as the most likely homolog for each contig. As an independent assessment of Kraken, we also ran all reads from the control samples through the CLC workflow without prior SNAP filtering/Kraken analysis.
The results of primary (Kraken) and secondary (CLC) analyses were screened for pathogens known to cause prominent symptoms in infected patients. Following this, the probable causative agents were listed according to the number of reads (from largest to smallest) obtained from the primary analysis. The BLAST e-value was reported if the pathogen was detected in the secondary analysis as well. When the sequencing was performed on duplicate samples, only the results from the sample that produced the largest sequencing dataset were reported. Finally, the results of bioinformatics analysis in conjunction with supporting clinical data and laboratory findings were used to inform diagnosis.
Results
Of the 40 DNA and cDNA samples that we prepared in duplicate, only 22 samples from 17 participants (eight samples from seven control subjects and 14 samples from ten test subjects) met the quantity and quality requirements for deep sequencing. From these, between 4 and 20 million reads per sample were generated. The majority (43.67–94.38 %) of these reads were of human origin, with only a small proportion of non-human reads being classified by Kraken. The number of viral, bacterial and archaeal species reported by Kraken varied considerably, from 146 to 505 species per sample (Fig. 3).
The secondary analysis facilitated further classification of reads left unclassified by Kraken. Analysis with the CLC Genomics Workbench revealed that Kraken-unclassified reads still contained human sequences, accounting for 18.9–81.7 % of the total contigs in each sample. Non-host contigs classified by BLAST analysis included viruses, bacteria and other organisms, such as archaea (e.g. Sulfolobus sp.), fungi (e.g. Saccharomyces sp., Cryptococcus sp., Penicillium sp.), algae (e.g. Navicula gregaria), plants (e.g. rice, tomato, grain, tobacco), protozoa (e.g. Toxoplasma gondii, Plasmodium berghei), human parasites (e.g. roundworm, tapeworm, pinworm) and larger animals (e.g. snail, fish, rat, monkey, orangutan, gorilla). Following analysis, 2.7–29.5 % of the total contigs in each sample remained unclassified (Fig. 4).
Summary of CLC Genomics analysis on Kraken-unclassified reads *BLASTn optimised for highly similar sequences (megablast) was performed to search the homology between query sequences and reference sequences in the database. If multiple significant similarities matched with a single species, only the highest scoring hit was reported. A similarity was considered significant at e-value ≤10−5
Table 1 compares the outcomes of hospital investigations and NGS analyses. Confirmation of diagnosis by NGS means that at least one read or one contig was assigned to the infectious organism. Kraken analysis confirmed the specific diagnoses obtained by other means in 7/8 (87.5 %) of control samples, whereas CLC analysis only concurred with the results of clinical tests in 2/8 (25 %) of control samples, both of whom had dengue virus 1 infection: patient ID# 005 and 017. The CLC reference mapping aligned the position of the virus contigs with the dengue virus 1 reference genome (accession number: NC_001477.1). In the first dengue case (ID# 005), the CLC analysis constructed eight contigs of dengue virus 1 with length 166 to 1328 bp, whereas in the second dengue case (ID# 017), only one contig of dengue virus 1 was available, with a length of 217 bp (Fig. 5).
Mapping of dengue virus 1 contigs against dengue virus 1 complete genome of 10,735 bp genomic DNA (NCBI Reference Sequence: NC_001477.1). The X-axis represents the size and position of the genome/contigs in base pair (bp); the Y-axis represents the dengue virus 1 reference genome and the dengue virus 1 contig(s) found in the sample ID# 5c and 17c1; both are cDNA samples from patient ID# 005 and 017, respectively
Our deep sequencing approach identified some plausible causes of fever in 80 % (8/10) of the test subjects (ID# 002, 011, 014, 019, 027, 029, 030, 039) who remained undiagnosed after routine hospital investiagations (Table 2). In particular, our analyses confirmed the aetiological diagnosis of two AUF cases that eluded conventional investigation methods. A high number of Escherichia coli sequences were detected in the sample of a fatal diarrhoea case with a sterile blood culture (patient ID# 002) and Orientia tsutsugamushi was detected in a PCR-negative patient with clinical features of scrub typhus (patient ID# 011). Furthermore, the results of deep sequencing highlighted a surprising microbial diversity in a ‘sterile’ environment; that is, human blood. Table 3 shows 61 organisms that were present in all samples and their relative abundance.
Discussion
To date, there has been little research undertaken to investigate the application of NGS in fever investigation, possibly due to the relatively high costs and complex methods of sample preparation and data analysis. Since the costs associated with NGS are proportional to the amount of sequence data generated per sequencing run, one way to reduce the sequencing costs is by requesting the minimum amount of sequence data without compromising the sensitivity of detection of the pathogen. This can be achieved, for instance, by reducing human DNA contamination to maximise the yield of pathogen sequences. Most human DNA is cellular in origin; thus, cell-free samples such as plasma and serum are expected to have a higher ratio of pathogen/human DNA than whole blood.
Previous studies have used deep sequencing to facilitate the discovery of novel viruses in plasma/serum samples [13, 18–21], but the use of such samples in the present study posed a challenge. Despite achieving low-level human DNA contamination, which is necessary to keep sequencing costs minimal, the plasma and serum samples contained very low quantity and quality of nucleic acids. Commercial sequencing services that we contacted required at least 100 ng of DNA/RNA per sample, which is challenging to achieve from plasma/serum volumes typically collected for routine blood diagnosis. The attempt to meet the minimum input requirement for sequencing necessitated amplification, which could introduce biases during sequencing [22]. Furthermore, every additional step carried out during sample preparation is a potential source of contamination, and might cause further degradation of the nucleic acids.
While the primary aim of this study was to evaluate the use of NGS in detecting pathogens associated with AUFs, our data provide an insight into the microbial diversity in human blood (Table 3). We presumed that the origins of these microorganisms are either from experimental reagents (i.e. Achromobacter xylosoxidans, Alteromonas macleodii, enterobacteria phage), cross-contamination from one sample to another (i.e. hepatitis C virus, dengue virus), from the skin during phlebotomy (i.e. Propionibacterium acnes, Staphylococcus epidermidis) or from the blood itself [i.e. torque teno midi virus (TTMDV), human herpesvirus 4].
Achromobacter xylosoxidans can be found in water environments and has been isolated from both immunocompetent and immunocompromised patients with bacteraemia, chronic otitis media, meningitis, urinary tract infections, abscesses, osteomyelitis, corneal ulcers, prosthetic valve endocarditis, peritonitis and pneumonia [23, 24]. The bacteria are not a typical component of human flora and have low virulence [25]. Infection with A. xylosoxidans is widely considered to be opportunistic, and the source of infection is usually found to be a contaminated solution [23]. The bacteria can survive in aqueous environments with minimal nutrients, so it is likely that the relatively high abundance of these bacteria (0.16–1.38 % of non-host reads) indicates contamination from the water or reagent used during sample preparation. It has been reported previously [26] that sequence-based microbiome analyses is susceptible to DNA contamination introduced by molecular biology grade water, PCR reagents and DNA extraction kits.
Alteromonas macleodii is commonly found in temperate or tropical sea waters [27, 28]. The presence of these Gram-negative bacteria in humans has not been reported. The present study detected high levels of A. macleodii reads across the samples, accounting for 0.51–11.35 % of non-host reads. It is suspected that this organism is a contaminant, and its presence in the NGS dataset should be disregarded.
Bacteriophage (phage) infects and replicates within a bacterium and can be present in the study samples through multiple routes. As Enterobacter is part of normal gut flora, there is obviously abundant enterobacteria phage in the human body. Assuming that the phage originated from the patients’ gut, the question is how this phage can escape the gut–blood barrier. On the other hand, sequencing of phage genomes is an interesting field of research, with potential uses for phages as antimicrobials and biocontrol agents for food production [29], and the phiX174 bacteriophage was the first DNA-based genome to be sequenced, dating back to 1977 [30]. A previous study [31] reported enterobacteria phage phiX174 sensu lato as a common contaminant in NGS datasets from blood samples.
The presence of hepatitis C virus and dengue virus in all samples is evidence of cross-contamination from one sample to another. One sample was collected from a patient with confirmed hepatitis C virus infection (patient ID# 006); however, this sample was not sequenced due to insufficient amounts of nucleic acids to enter the amplification step. Accordingly, it was presumed that the low abundance (<0.1 %) of hepatitis C virus reads in all samples was the result of contamination during DNA/RNA isolation. As for dengue virus, although it was detected in all samples, its presence with high read counts (2.58 %) in sample ID# 5c indicates a true infection. Dengue virus reads were also present in a relatively higher proportion (0.01 %) in sample ID# 14 compared to the rest of the samples. It is possible that patient ID# 014 had a dengue infection.
Human herpesvirus 4 or Epstein–Barr virus (EBV) is one of the most common viruses in humans. This virus is widespread internationally, and around 95 % of the human population is infected with EBV [32]. Therefore, the presence of low levels (<1 % of non-host reads) of EBV in all samples is not surprising. TTMDV has been found in various body fluids, including saliva and nasopharyngeal aspirates, serum, urine and stool collected from children with acute respiratory disease [33]. The frequent detection of TTMDV in our study is consistent with a previous metagenomic study [34], which reported that TTMDV constituted the second largest viral community after torque teno virus in the plasma of healthy adults.
Although most of the organisms detected in the samples were presumably contaminants, our data illustrate the high sensitivity of the deep sequencing approach to reveal microbial diversity within a sample. Previous research [35] had successfully detected bacterial 16S ribosomal DNA sequences that were similar to DNA sequences of Riemerella anatipestifer, Pseudomonas fluorescens, Propionibacterium acnes, Microbacterium schleiferi, Stenotrophomonas and Pseudomonas putida in healthy human blood using real-time PCR and traditional sequencing on an ABI PRISM platform. These bacterial sequences presumably originated either from experimental reagents, from the skin during phlebotomy or from the blood itself. Nonetheless, the findings of the study raised the possibility that there is a ‘normal’ population of bacterial DNA sequences in blood that has previously been considered sterile. The use of NGS in the present study facilitated the identification of organisms that might well escape cultivation because of their low burden in the blood or simply because they are unculturable. With only 1 % of the microbial life on earth able to be cultured [36, 37], culture-independent methods, such as deep sequencing, are clearly required in order to extend our knowledge of microbial diversity.
In addition to revealing microbial diversity in human blood, this study provided important information with regards to the sensitivity and cost-effectiveness of the deep sequencing approach for fever investigation. We demonstrated that, with an optimum sample such as that from patient ID# 005, approximately 15 million PE reads (~2 Gb of data) are sufficient to facilitate the diagnosis of dengue virus. When the virus load was sufficiently high, de novo assembly permitted the generation of several contigs corresponding to a nearly full-length genome, thus confirming the diagnosis of dengue fever at a cost of AUD $600 for sample processing, library preparation and deep sequencing.
Bioinformatics analyses are an enormous challenge in metagenomic studies because, while a variety of tools are available for analysing sequence data, they require expert users to assemble them into an effective workflow. The high volume of data generated by NGS technology is often responsible for considerable delays in achieving a robust diagnosis. In our experience, bioinformatics analyses of ~2 Gb of raw sequence data using Kraken can be completed in 1 h. This speed far exceeds that of conventional BLAST searches, which may take more than 24 h to complete. To complete the data analysis as quickly as possible, we used the CLC Genomic Workbench workflow on reads unclassified by Kraken to facilitate the identification of infectious agents in undiagnosed cases (test subjects). In order to validate Kraken findings, we performed CLC analysis on the raw reads originating from control subjects in parallel with Kraken analysis. As Kraken reported an excessive amount (hundreds) of organisms in a single sample, the usefulness of Kraken in the present study relates to the rapid screening of pathogens associated with fever. There is a possibility that ambiguous reads may be mapped to multiple taxa in the MiniKraken database, resulting in the detection of organisms that were not actually present in the sample (false-positive). We suggest that, if the Kraken program is going to be used for diagnosis, additional steps are required to filter the true pathogen or pathogen sequences with clinical relevance. Most importantly, the Kraken findings are best interpreted in conjunction with reliable clinical information and the results of other tests are absolutely necessary to inform diagnosis.
We showed in Table 1 that performing more complex analyses does not guarantee reliability of results, and may, indeed, result in failure to detect the true cause of fever (false-negative). The CLC analysis workflow, which involved pre-processing and assembly prior to the BLAST search, failed to detect the true pathogen in 75 % (6/8) of control samples. Thus, it can be argued that the success of the deep sequencing approach is more likely to be determined by the sample condition and the sequencing dataset rather than the analysis pipeline. It is important to collect samples with high pathogen load. Ideally, samples should be snap frozen in liquid nitrogen to preserve the scarce amount of nucleic acids from the pathogen. It is equally important to keep the sample processing steps to a minimum to avoid contamination and prevent further degradation of the nucleic acids. Providing negative controls and running ‘blanks’ in the same sequencing lanes as the actual samples can make the interpretation of the results easier, as the contaminating agents can be immediately identified and ruled out from the NGS dataset. Finally, the choice of NGS platform contributes to the success of pathogen detection. The long reads produced by the 454 platform increase the specificity of pathogen identification by facilitating the discrimination of pathogen reads from hosts or endogenous flora. Although Illumina platforms generate short reads (currently up to 2 × 150 bp for the HiSeq and Genome Analyzer II and 2 × 300 bp for the MiSeq), this platform can generate sequencing data much faster than the 454 platform, thus providing sufficient read depth or number of sequence reads generated per run to detect pathogens with a high degree of sensitivity.
The development of single-molecule sequencing (also known as third-generation sequencing), such as nanopore sequencing or MinION (Oxford Nanopore Technologies), is highly promising; this pocket-sized genome sequencer can generate longer reads (tens of kilobases) at a cost comparable to that of currently available NGS instruments [38]. Another advantage of this portable DNA sequencer over NGS is its ability to perform real-time sequence analysis, which is highly valuable for providing results rapidly. The downside of nanopore sequencing is that the technology currently requires micrograms of DNA/cDNA input. Two recent studies [39, 40] reported disadvantages of MinION sequencing, including higher error rates (10–30 %) and relatively lower throughput (<100,000 reads per cell) compared with NGS.
In conclusion, the deep sequencing approach facilitated the identification of infectious agents associated with AUF as well as other organisms present in human blood. We identified challenges in conducting a deep sequencing approach for routine investigation of fever. Future improvements in sequencing platforms are needed to provide longer reads and enable sequencing from smaller amounts of input material. The development of bioinformatics tools should be directed towards user-friendly options and the means to provide answers in clinically relevant timeframes (e.g. within hours of sample receipt). Recent advancement in sequencing technologies and bioinformatics analyses provides a positive outlook for the application of the deep sequencing approach to facilitate the diagnosis of AUFs.
Abbreviations
- AGRF:
-
Australian Genome Research Facility
- ANA:
-
Antinuclear antibody
- ALT:
-
Alanine aminotransferase
- AST:
-
Aspartate aminotransferase
- AUF:
-
Acute undifferentiated fever
- BLAST:
-
Basic Local Alignment Search Tool
- CMV:
-
Cytomegalovirus
- CRP:
-
C-reactive protein
- CT:
-
Computed tomography
- DNA:
-
Deoxyribonucleic acid
- EBV:
-
Epstein–Barr virus
- NCBI:
-
National Center for Biotechnology Information
- NGS:
-
Next-generation sequencing
- PCR:
-
Polymerase chain reaction
- RNA:
-
Ribonucleic acid
- SFG:
-
Spotted fever group
- TTMDV:
-
Torque teno midi virus
- WBC:
-
White blood cell
References
Susilawati TN, McBride WJ (2014) Acute undifferentiated fever in Asia: a review of the literature. Southeast Asian J Trop Med Public Health 45(3):719–726
Cunha BA (2007) Fever of unknown origin: focused diagnostic approach based on clinical clues from the history, physical examination, and laboratory tests. Infect Dis Clin North Am 21(4):1137–1187. doi:10.1016/j.idc.2007.09.004, xi
Abrahamsen SK, Haugen CN, Rupali P, Mathai D, Langeland N, Eide GE, Mørch K (2013) Fever in the tropics: aetiology and case-fatality—a prospective observational study in a tertiary care hospital in South India. BMC Infect Dis 13:355. doi:10.1186/1471-2334-13-355
Sint D, Raso L, Traugott M (2012) Advances in multiplex PCR: balancing primer efficiencies and improving detection success. Methods Ecol Evol 3(5):898–905. doi:10.1111/j.2041-210X.2012.00215.x
Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE (2000) Multiplex PCR: optimization and application in diagnostic virology. Clin Microbiol Rev 13(4):559–570
Hsu CC, Tokarz R, Briese T, Tsai HC, Quan PL, Lipkin WI (2013) Use of staged molecular analysis to determine causes of unexplained central nervous system infections. Emerg Infect Dis 19(9):1470–1477. doi:10.3201/eid1909.130474
McLoughlin KS (2011) Microarrays for pathogen detection and analysis. Brief Funct Genomics 10(6):342–353. doi:10.1093/bfgp/elr027
Dunbar SA, Ritchie VB, Hoffmeyer MR, Rana GS, Zhang H (2015) Luminex(®) multiplex bead suspension arrays for the detection and serotyping of Salmonella spp. Methods Mol Biol 1225:1–27. doi:10.1007/978-1-4939-1625-2_1
Yozwiak NL, Skewes-Cox P, Stenglein MD, Balmaseda A, Harris E, DeRisi JL (2012) Virus identification in unknown tropical febrile illness cases using deep sequencing. PLoS Negl Trop Dis 6(2):e1485. doi:10.1371/journal.pntd.0001485
Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M (2012) Comparison of next-generation sequencing systems. J Biomed Biotechnol 2012:251364. doi:10.1155/2012/251364
Calistri A, Palù G (2015) Editorial commentary: Unbiased next-generation sequencing and new pathogen discovery: undeniable advantages and still-existing drawbacks. Clin Infect Dis 60(6):889–891. doi:10.1093/cid/ciu913
Perlejewski K, Popiel M, Laskus T, Nakamura S, Motooka D, Stokowy T, Lipowski D, Pollak A, Lechowicz U, Caraballo Cortés K, Stępień A, Radkowski M, Bukowska-Ośko I (2015) Next-generation sequencing (NGS) in the identification of encephalitis-causing viruses: unexpected detection of human herpesvirus 1 while searching for RNA pathogens. J Virol Methods 226:1–6. doi:10.1016/j.jviromet.2015.09.010
Grard G, Fair JN, Lee D, Slikas E, Steffen I, Muyembe JJ, Sittler T, Veeraraghavan N, Ruby JG, Wang C, Makuwa M, Mulembakani P, Tesh RB, Mazet J, Rimoin AW, Taylor T, Schneider BS, Simmons G, Delwart E, Wolfe ND, Chiu CY, Leroy EM (2012) A novel rhabdovirus associated with acute hemorrhagic fever in central Africa. PLoS Pathog 8(9):e1002924. doi:10.1371/journal.ppat.1002924
Zaharia M, Bolosky WJ, Curtis K, Fox A, Patterson D, Shenker S, Stoica I, Karp RM, Sittler T (2011) Faster and more accurate sequence alignment with SNAP. arXiv arXiv (1111.5572v1)
Wood DE, Salzberg SL (2014) Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15(3):R46. doi:10.1186/gb-2014-15-3-r46
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Home page at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 3 November 2015
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. doi:10.1016/s0022-2836(05)80360-2
Palacios G, Druce J, Du L, Tran T, Birch C, Briese T, Conlan S, Quan PL, Hui J, Marshall J, Simons JF, Egholm M, Paddock CD, Shieh WJ, Goldsmith CS, Zaki SR, Catton M, Lipkin WI (2008) A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med 358(10):991–998. doi:10.1056/NEJMoa073785
Briese T, Paweska JT, McMullan LK, Hutchison SK, Street C, Palacios G, Khristova ML, Weyer J, Swanepoel R, Egholm M, Nichol ST, Lipkin WI (2009) Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog 5(5):e1000455. doi:10.1371/journal.ppat.1000455
Xu B, Liu L, Huang X, Ma H, Zhang Y, Du Y, Wang P, Tang X, Wang H, Kang K, Zhang S, Zhao G, Wu W, Yang Y, Chen H, Mu F, Chen W (2011) Metagenomic analysis of fever, thrombocytopenia and leukopenia syndrome (FTLS) in Henan Province, China: discovery of a new bunyavirus. PLoS Pathog 7(11):e1002369. doi:10.1371/journal.ppat.1002369
McMullan LK, Folk SM, Kelly AJ, MacNeil A, Goldsmith CS, Metcalfe MG, Batten BC, Albariño CG, Zaki SR, Rollin PE, Nicholson WL, Nichol ST (2012) A new phlebovirus associated with severe febrile illness in Missouri. N Engl J Med 367(9):834–841. doi:10.1056/NEJMoa1203378
Pinard R, de Winter A, Sarkis GJ, Gerstein MB, Tartaro KR, Plant RN, Egholm M, Rothberg JM, Leamon JH (2006) Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genomics 7:216. doi:10.1186/1471-2164-7-216
Duggan JM, Goldstein SJ, Chenoweth CE, Kauffman CA, Bradley S (1996) Achromobacter xylosoxidans bacteremia: report of four cases and review of the literature. Clin Infect Dis 23(3):569–576. doi:10.1093/clinids/23.3.569
Igra-Siegman Y, Chmel H, Cobbs C (1980) Clinical and laboratory characteristics of Achromobacter xylosoxidans infection. J Clin Microbiol 11(2):141–145
Claassen SL, Reese JM, Mysliwiec V, Mahlen SD (2011) Achromobacter xylosoxidans infection presenting as a pulmonary nodule mimicking cancer. J Clin Microbiol 49(7):2751–2754. doi:10.1128/JCM.02571-10
Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW (2014) Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. doi:10.1186/s12915-014-0087-z
López-Pérez M, Gonzaga A, Martin-Cuadrado A-B, Onyshchenko O, Ghavidel A, Ghai R, Rodriguez-Valera F (2012) Genomes of surface isolates of Alteromonas macleodii: the life of a widespread marine opportunistic copiotroph. Sci Rep 2:696. doi:10.1038/srep00696
Ivars-Martinez E, Martin-Cuadrado AB, D’Auria G, Mira A, Ferriera S, Johnson J, Friedman R, Rodriguez-Valera F (2008) Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. ISME J 2(12):1194–1212. doi:10.1038/ismej.2008.74
Klumpp J, Fouts DE, Sozhamannan S (2012) Next generation sequencing technologies and the changing landscape of phage genomics. Bacteriophage 2(3):190–199. doi:10.4161/bact.22111
Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M (1977) Nucleotide sequence of bacteriophage phi X174 DNA. Nature 265(5596):687–695
Lei H, Li T, Hung GC, Li B, Tsai S, Lo SC (2013) Identification and characterization of EBV genomes in spontaneously immortalized human peripheral blood B lymphocytes by NGS technology. BMC Genomics 14:804. doi:10.1186/1471-2164-14-804
Walling DM, Shebib N, Weaver SC, Nichols CM, Flaitz CM, Webster-Cyriaque J (1999) The molecular epidemiology and evolution of Epstein-Barr virus: sequence variation and genetic recombination in the latent membrane protein-1 gene. J Infect Dis 179(4):763–774. doi:10.1086/314672
Burián Z, Szabó H, Székely G, Gyurkovits K, Pankovics P, Farkas T, Reuter G (2011) Detection and follow-up of torque teno midi virus (“small anelloviruses”) in nasopharyngeal aspirates and three other human body fluids in children. Arch Virol 156(9):1537–1541. doi:10.1007/s00705-011-1021-0
Li SK, Leung RKK, Guo HX, Wei JF, Wang JH, Kwong KT, Lee SS, Zhang C, Tsui SKW (2012) Detection and identification of plasma bacterial and viral elements in HIV/AIDS patients in comparison to healthy adults. Clin Microbiol Infect 18(11):1126–1133. doi:10.1111/j.1469-0691.2011.03690.x
Nikkari S, McLaughlin IJ, Bi W, Dodge DE, Relman DA (2001) Does blood of healthy subjects contain bacterial ribosomal DNA? J Clin Microbiol 39(5):1956–1959
Hugon P, Dufour JC, Colson P, Fournier PE, Sallah K, Raoult D (2015) A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect Dis 15(10):1211–1219
Vartoukian SR, Palmer RM, Wade WG (2010) Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol Lett 309(1):1–7
Laver T, Harrison J, O’Neill PA, Moore K, Farbos A, Paszkiewicz K, Studholme DJ (2015) Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol Detect Quantif 3:1–8. doi:10.1016/j.bdq.2015.02.001
Ashton PM, Nair S, Dallman T, Rubino S, Rabsch W, Mwaigwisya S, Wain J, O’Grady J (2015) MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol 33(3):296–300. doi:10.1038/nbt.3103
Kilianski A, Haas JL, Corriveau EJ, Liem AT, Willis KL, Kadavy DR, Rosenzweig CN, Minot SS (2015) Bacterial and viral identification and differentiation by amplicon sequencing on the MinION nanopore sequencer. Gigascience 4:12. doi:10.1186/s13742-015-0051-z
Acknowledgements
The authors wish to thank Sue Richmond for her assistance with the collection of blood samples from the study participants and Dr. Paul Giacomin, Dr. Annette Dougall and Mr. Darren Pickering for their intellectual contribution and assistance during sample preparation for sequencing. Financial support for this work was provided by James Cook University and Far North Queensland Hospital Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Susilawati, T.N., Jex, A.R., Cantacessi, C. et al. Deep sequencing approach for investigating infectious agents causing fever. Eur J Clin Microbiol Infect Dis 35, 1137–1149 (2016). https://doi.org/10.1007/s10096-016-2644-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-016-2644-6