Abstract
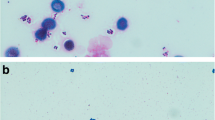
Blood is one of the most important specimens sent to a microbiology laboratory for culture. Most blood cultures are incubated for 5–7 days, except in cases where there is a suspicion of infection caused by microorganisms that proliferate slowly, or infections expressed by a small number of bacteria in the bloodstream. Therefore, at the end of incubation, misidentification of positive cultures and false-negative results are a real possibility. The aim of this work was to perform a confirmation by Gram staining of the lack of any microorganisms in blood cultures that were identified as negative by the BACTEC™ FX system at the end of incubation. All bottles defined as negative by the BACTEC FX system were Gram-stained using an automatic device and inoculated on solid growth media. In our work, 15 cultures that were defined as negative by the BACTEC FX system at the end of the incubation were found to contain microorganisms when Gram-stained. The main characteristic of most bacteria and fungi growing in the culture bottles that were defined as negative was slow growth. This finding raises a problematic issue concerning the need to perform Gram staining of all blood cultures, which could overload the routine laboratory work, especially laboratories serving large medical centers and receiving a large number of blood cultures.
Similar content being viewed by others
References
Magadia RR, Weinstein MP (2001) Laboratory diagnosis of bacteremia and fungemia. Infect Dis Clin North Am 15:1009–1024
Mylotte JM, Tayara A (2000) Blood cultures: clinical aspects and controversies. Eur J Clin Microbiol Infect Dis 19(3):157–163
Roh KH, Kim JY, Kim HN, Lee HJ, Sohn JW, Kim MJ, Cho Y, Kim YK, Lee CK (2012) Evaluation of BACTEC Plus aerobic and anaerobic blood culture bottles and BacT/Alert FAN aerobic and anaerobic blood culture bottles for the detection of bacteremia in ICU patients. Diagn Microbiol Infect Dis 73(3):239–242
Wang Y, Shi LN, Fan M, Huang M, Wang WP, Xi HY, Hu YA, Jia X, Shao HF (2012) Comparison of ability and characteristics of Bactec FX and Bact/Alert 3D blood culture systems in detection of bacteremia. Chin J Clin Lab Sci 1:006
McDonald CP, Rogers A, Cox M, Smith R, Roy A, Robbins S, Hartley S, Barbara JA, Rothenberg S, Stutzman L, Widders G (2002) Evaluation of the 3D BacT/ALERT automated culture system for the detection of microbial contamination of platelet concentrates. Transfus Med 12(5):303–309
Forbes BA, Sahm DF, Weissfeld AS (2007) Bailey & Scott’s diagnostic microbiology, 12th edn. Mosby Elsevier, Maryland Heights, pp 788–798
Klaerner H-G, Eschenbach U, Kamereck K, Lehn N, Wagner H, Miethke T (2000) Failure of an automated blood culture system to detect nonfermentative gram-negative bacteria. J Clin Microbiol 38(3):1036–1041
Houpikian P, Raoult D (2005) Blood culture-negative endocarditis in a reference center: etiologic diagnosis of 348 cases. Medicine (Baltimore) 84(3):162–173
Friedman ND, Kaye KS, Stout JE, McGarry SA, Trivette SL, Briggs JP, Lamm W, Clark C, MacFarquhar J, Walton AL, Reller LB, Sexton DJ (2002) Health care-associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med 137(10):791–797
Wenzel RP, Edmond MB (2001) The impact of hospital-acquired bloodstream infections. Emerg Infect Dis 7(2):174–177
Peters RP, van Agtmael MA, Danner SA, Savelkoul PH, Vandenbroucke-Grauls CM (2004) New developments in the diagnosis of bloodstream infections. Lancet Infect Dis 4(12):751–760
Felek S, Arslan A (1999) Gram staining with an automatic machine. Folia Microbiol (Praha) 44(3):333–337
Pugliese A, Pacris B, Schoch PE, Cunha BA (1993) Oligella urethralis urosepsis. Clin Infect Dis 17(6):1069–1070
Selvarangan R, Bui U, Limaye AP, Cookson BT (2003) Rapid identification of commonly encountered Candida species directly from blood culture bottles. J Clin Microbiol 41(12):5660–5664
Compliance with ethical standards
ᅟ
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Peretz, A., Isakovich, N., Pastukh, N. et al. Performance of Gram staining on blood cultures flagged negative by an automated blood culture system. Eur J Clin Microbiol Infect Dis 34, 1539–1541 (2015). https://doi.org/10.1007/s10096-015-2383-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-015-2383-0