Abstract
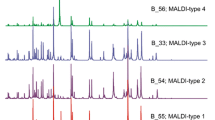
Reliable, quick and low-cost methods are needed for the early detection of multidrug-resistant and highly virulent high-risk B2 and D Escherichia coli clones or clonal complexes (HiRCC). Matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS) seems to have a good discriminatory potential at different subspecies levels, but it was never evaluated for the discrimination of E. coli clones. We assessed the potential of MALDI-TOF MS coupled to multivariate data analysis to discriminate representative E. coli B2 and D HiRCC. Seventy-three E. coli isolates from B2 (including ST131 and B2 non-ST131 clones) and D (ST69, ST393, ST405) with variable pulsed-field gel electrophoresis (PFGE) patterns, origins and dates (1980–2010) were tested. MS spectra were acquired from independent extracts obtained from different plate cultures in two different Microflex LT MALDI-TOF devices (Bruker) after a standard extraction procedure. MALDI-TOF MS fingerprinting analysis revealed a good discriminatory ability between the four HiRCC analysed (ST131, ST69, ST405, ST393) and between B2 ST131 and other B2 non-ST131 isolates. Clusters defined by MALDI-TOF MS were consistent with the clonal complexes assigned by multilocus sequence typing (MLST), although differences were detected regarding the composition of clusters obtained by the comparison of PFGE profiles. We demonstrate, for the first time, that characteristic mass fingerprints of different E. coli HiRCC are sufficiently discriminatory and robust to enable their differentiation by MALDI-TOF MS, which might represent a promising tool for the optimisation of infection control, individual patient management and large-scale epidemiological studies of public health relevance. The good correlation between phenotypic and genotypic features further corroborates phylogenetic relationships delineated by MLST.




Similar content being viewed by others
References
Baquero F, Coque TM (2011) Multilevel population genetics in antibiotic resistance. FEMS Microbiol Rev 35(5):705–706
Woodford N, Turton JF, Livermore DM (2011) Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35(5):736–755
Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M (2010) Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis 51(3):286–294
Johnson JR, Tchesnokova V, Johnston B, Clabots C, Roberts PL, Billig M, Riddell K, Rogers P, Qin X, Butler-Wu S, Price LB, Aziz M, Nicolas-Chanoine MH, Debroy C, Robicsek A, Hansen G, Urban C, Platell J, Trott DJ, Zhanel G, Weissman SJ, Cookson BT, Fang FC, Limaye AP, Scholes D, Chattopadhyay S, Hooper DC, Sokurenko EV (2013) Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J Infect Dis 207(6):919–928
Centers for Disease Control and Prevention (CDC) (2013) Antibiotic resistance threats in the United States, 2013. Available online at: http://www.cdc.gov/drugresistance/threat-report-2013/. Last accessed 13 December 2013
Novais A, Pires J, Ferreira H, Costa L, Montenegro C, Vuotto C, Donelli G, Coque TM, Peixe L (2012) Characterization of globally spread Escherichia coli ST131 isolates (1991 to 2010). Antimicrob Agents Chemother 56(7):3973–3976
Novais Â, Vuotto C, Pires J, Montenegro C, Donelli G, Coque TM, Peixe L (2013) Diversity and biofilm-production ability among isolates of Escherichia coli phylogroup D belonging to ST69, ST393 and ST405 clonal groups. BMC Microbiol 13:144
Blanco J, Mora A, Mamani R, López C, Blanco M, Dahbi G, Herrera A, Blanco JE, Alonso MP, García-Garrote F, Chaves F, Orellana MÁ, Martínez-Martínez L, Calvo J, Prats G, Larrosa MN, González-López JJ, López-Cerero L, Rodríguez-Baño J, Pascual A (2011) National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain. J Antimicrob Chemother 66(9):2011–2021
Blanco J, Mora A, Mamani R, López C, Blanco M, Dahbi G, Herrera A, Marzoa J, Fernández V, de la Cruz F, Martínez-Martínez L, Alonso MP, Nicolas-Chanoine MH, Johnson JR, Johnston B, López-Cerero L, Pascual A, Rodríguez-Baño J; Spanish Group for Nosocomial Infections (GEIH) (2013) Four main virotypes among extended-spectrum-beta-lactamase-producing isolates of Escherichia coli O25b:H4-B2-ST131: bacterial, epidemiological, and clinical characteristics. J Clin Microbiol 51(10):3358–3367
Dahbi G, Mora A, López C, Alonso MP, Mamani R, Marzoa J, Coira A, García-Garrote F, Pita JM, Velasco D, Herrera A, Viso S, Blanco JE, Blanco M, Blanco J (2013) Emergence of new variants of ST131 clonal group among extraintestinal pathogenic Escherichia coli producing extended-spectrum beta-lactamases. Int J Antimicrob Agents 42(4):347–351
Clark AE, Kaleta EJ, Arora A, Wolk DM (2013) Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev 26(3):547–603
Fenselau C, Demirev PA (2001) Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom Rev 20(4):157–171
Lartigue MF (2013) Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for bacterial strain characterization. Infect Genet Evol 13:230–235
Clark CG, Kruczkiewicz P, Guan C, McCorrister SJ, Chong P, Wylie J, van Caeseele P, Tabor HA, Snarr P, Gilmour MW, Taboada EN, Westmacott GR (2013) Evaluation of MALDI-TOF mass spectroscopy methods for determination of Escherichia coli pathotypes. J Microbiol Methods 94(3):180–191
Hrabák J, Chudácková E, Walková R (2013) Matrix-assisted laser desorption ionization-time of flight (maldi-tof) mass spectrometry for detection of antibiotic resistance mechanisms: from research to routine diagnosis. Clin Microbiol Rev 26(1):103–114
Momo RA, Povey JF, Smales CM, O’Malley CJ, Montague GA, Martin EB (2013) MALDI-ToF mass spectrometry coupled with multivariate pattern recognition analysis for the rapid biomarker profiling of Escherichia coli in different growth phases. Anal Bioanal Chem 405(25):8251–8265
Kuehl B, Marten SM, Bischoff Y, Brenner-Weiss G, Obst U (2011) MALDI-ToF mass spectrometry-multivariate data analysis as a tool for classification of reactivation and non-culturable states of bacteria. Anal Bioanal Chem 401(5):1593–1600
Dieckmann R, Malorny B (2011) Rapid screening of epidemiologically important Salmonella enterica subsp. enterica serovars by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl Environ Microbiol 77(12):4136–4146
Barbuddhe SB, Maier T, Schwarz G, Kostrzewa M, Hof H, Domann E, Chakraborty T, Hain T (2008) Rapid identification and typing of Listeria species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl Environ Microbiol 74(17):5402–5407
Wolters M, Rohde H, Maier T, Belmar-Campos C, Franke G, Scherpe S, Aepfelbacher M, Christner M (2011) MALDI-TOF MS fingerprinting allows for discrimination of major methicillin-resistant Staphylococcus aureus lineages. Int J Med Microbiol 301(1):64–68
Böhme K, Fernández-No IC, Pazos M, Gallardo JM, Barros-Velázquez J, Cañas B, Calo-Mata P (2013) Identification and classification of seafood-borne pathogenic and spoilage bacteria: 16S rRNA sequencing versus MALDI-TOF MS fingerprinting. Electrophoresis 34(6):877–887
Bernaschi P, Del Chierico F, Petrucca A, Argentieri A, Ciofi Degli Atti M, Ciliento G, Carletti M, Muraca M, Locatelli F, Putignani L (2013) Microbial tracking of multidrug-resistant Klebsiella pneumoniae isolates in a pediatric hospital setting. Int J Immunopathol Pharmacol 26(2):463–472
Berrazeg M, Diene SM, Drissi M, Kempf M, Richet H, Landraud L, Rolain JM (2013) Biotyping of multidrug-resistant Klebsiella pneumoniae clinical isolates from France and Algeria using MALDI-TOF MS. PLoS One 8(4):e61428
Welker M (2011) Proteomics for routine identification of microorganisms. Proteomics 11(15):3143–3153
Croxatto A, Prod’hom G, Greub G (2012) Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol Rev 36(2):380–407
Vlek AL, Bonten MJ, Boel CH (2012) Direct matrix-assisted laser desorption ionization time-of-flight mass spectrometry improves appropriateness of antibiotic treatment of bacteremia. PLoS One 7(3):e32589
Del Chierico F, Petrucca A, Vernocchi P, Bracaglia G, Fiscarelli E, Bernaschi P, Muraca M, Urbani A, Putignani L (2014) Proteomics boosts translational and clinical microbiology. J Proteomics 97:69–87
Clerc O, Prod’hom G, Vogne C, Bizzini A, Calandra T, Greub G (2013) Impact of matrix-assisted laser desorption ionization time-of-flight mass spectrometry on the clinical management of patients with Gram-negative bacteremia: a prospective observational study. Clin Infect Dis 56(8):1101–1107
Naes T, Isaksson T, Fearn T, Davis T (2002) A user-friendly guide to multivariate calibration and classification. NIR Publications, Chichester
Geladi P, Kowalsky BR (1986) Partial least-squares regression: a tutorial. Anal Chim Acta 185:1–17
Alsberg BK, Kell DB, Goodacre R (1998) Variable selection in discriminant partial least-squares analysis. Anal Chem 70:4126–4133
De Maesschalck R, Candolfi A, Massart DL, Heuerding S (1999) Decision criteria for soft independent modeling of class analogy applied to near infrared data. Chemom Intell Lab Syst 47:65–77
Wold S (1976) Pattern recognition by means of disjoint principal components models. Pattern Recog 8:127–139
Hunter PR (1990) Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol 28(9):1903–1905
Josten M, Reif M, Szekat C, Al-Sabti N, Roemer T, Sparbier K, Kostrzewa M, Rohde H, Sahl HG, Bierbaum G (2013) Analysis of the matrix-assisted laser desorption ionization-time of flight mass spectrum of Staphylococcus aureus identifies mutations that allow differentiation of the main clonal lineages. J Clin Microbiol 51(6):1809–1817
Sandrin TR, Goldstein JE, Schumaker S (2013) MALDI TOF MS profiling of bacteria at the strain level: a review. Mass Spectrom Rev 32(3):188–217
Lasch P, Drevinek M, Nattermann H, Grunow R, Stämmler M, Dieckmann R, Schwecke T, Naumann D (2010) Characterization of Yersinia using MALDI-TOF mass spectrometry and chemometrics. Anal Chem 82(20):8464–8475
Dworzanski JP, Deshpande SV, Chen R, Jabbour RE, Snyder AP, Wick CH, Li L (2006) Mass spectrometry-based proteomics combined with bioinformatic tools for bacterial classification. J Proteome Res 5(1):76–87
Williams TL, Andrzejewski D, Lay JO, Musser SM (2003) Experimental factors affecting the quality and reproducibility of MALDI TOF mass spectra obtained from whole bacteria cells. J Am Soc Mass Spectrom 14(4):342–351
Dieckmann R, Helmuth R, Erhard M, Malorny B (2008) Rapid classification and identification of salmonellae at the species and subspecies levels by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl Environ Microbiol 74(24):7767–7778
Mellmann A, Bimet F, Bizet C, Borovskaya AD, Drake RR, Eigner U, Fahr AM, He Y, Ilina EN, Kostrzewa M, Maier T, Mancinelli L, Moussaoui W, Prévost G, Putignani L, Seachord CL, Tang YW, Harmsen D (2009) High interlaboratory reproducibility of matrix-assisted laser desorption ionization-time of flight mass spectrometry-based species identification of nonfermenting bacteria. J Clin Microbiol 47(11):3732–3734
Wunschel SC, Jarman KH, Petersen CE, Valentine NB, Wahl KL, Schauki D, Jackman J, Nelson CP, White E 5th (2005) Bacterial analysis by MALDI-TOF mass spectrometry: an inter-laboratory comparison. J Am Soc Mass Spectrom 16(4):456–462
Suarez S, Ferroni A, Lotz A, Jolley KA, Guérin P, Leto J, Dauphin B, Jamet A, Maiden MC, Nassif X, Armengaud J (2013) Ribosomal proteins as biomarkers for bacterial identification by mass spectrometry in the clinical microbiology laboratory. J Microbiol Methods 94(3):390–396
Rodrigues C, Machado E, Peixe L, Novais A (2013) IncI1/ST3 and IncN/ST1 plasmids drive the spread of bla TEM-52 and bla CTX-M-1/-32 in diverse Escherichia coli clones from different piggeries. J Antimicrob Chemother 68(10):2245–2248
Acknowledgements
This work was supported by FEDER funds through the Programa Operacional Factores de Competitividade—COMPETE and by National Funds through Fundação para a Ciência e a Tecnologia (FCT) through grant numbers EXPL/DTP-EPI/0196/2012 and FCOMP-01-0124-FEDER-027745, and also PEst-C/EQB/LA0006/2013, and an ESCMID research grant. The work received also inancial support from the European Union (FEDER funds) under the framework of QREN through Project NORTE-07-0124-FEDER-000066. AN was supported by a Marie Curie Intra-European Fellowship within the 7th European Community Framework Programme (PIEF-GA-2009-255512) and CS was supported by a post-doctoral grant (SFRH/BPD/70548/2010). Work at the Hospital Ramón y Cajal (RC and TMC’s groups) is funded by grants from the European Union (EVOTAR-LSHM-2011-282004 and R-GNOSIS-LHSM-2011-282512), the Ministry of Economy and Competitiveness—ISCIII of Spain (PI12/01581, REIPI-RD12/0015) and the regional government of Madrid (S2010/BMD2414_PROMPT-CM).
Conflict of interest
Rafael Cantón has participated in educational programmes organised by Bruker and in consultancy activities organised by Thermo Fisher.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Novais, Â., Sousa, C., de Dios Caballero, J. et al. MALDI-TOF mass spectrometry as a tool for the discrimination of high-risk Escherichia coli clones from phylogenetic groups B2 (ST131) and D (ST69, ST405, ST393). Eur J Clin Microbiol Infect Dis 33, 1391–1399 (2014). https://doi.org/10.1007/s10096-014-2071-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-014-2071-5