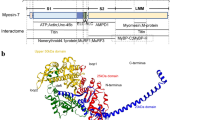
Abstract
Mitochondrial myopathies belong to a larger group of systemic diseases caused by morphological or biochemical abnormalities of mitochondria. Mitochondrial disorders can be caused by mutations in either the mitochondrial or nuclear genome. Only 5 % of all mitochondrial disorders are autosomal dominant. We analyzed DNA from members of the previously reported Puerto Rican kindred with an autosomal dominant mitochondrial myopathy (Heimann-Patterson et al. 1997). Linkage analysis suggested a putative locus on the pericentric region of the long arm of chromosome 22 (22q11). Using the tools of integrative genomics, we established chromosome 22 open reading frame 16 (C22orf16) (later designated as CHCHD10) as the only high-scoring mitochondrial candidate gene in our minimal candidate region. Sequence analysis revealed a double-missense mutation (R15S and G58R) in cis in CHCHD10 which encodes a coiled coil-helix-coiled coil-helix protein of unknown function. These two mutations completely co-segregated with the disease phenotype and were absent in 1,481 Caucasian and 80 Hispanic (including 32 Puerto Rican) controls. Expression profiling showed that CHCHD10 is enriched in skeletal muscle. Mitochondrial localization of the CHCHD10 protein was confirmed using immunofluorescence in cells expressing either wild-type or mutant CHCHD10. We found that the expression of the G58R, but not the R15S, mutation induced mitochondrial fragmentation. Our findings identify a novel gene causing mitochondrial myopathy, thereby expanding the spectrum of mitochondrial myopathies caused by nuclear genes. Our findings also suggest a role for CHCHD10 in the morphologic remodeling of the mitochondria.






Similar content being viewed by others
References
Debray FG, Lambert M, Mitchell GA (2008) Disorders of mitochondrial function. Curr Opin Pediatr 20:471–482
Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F et al (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465
Lopez MF, Kristal BS, Chernokalskaya E, Lazarev A, Shestopalov AI, Bogdanova A, Robinson M (2000) High-throughput profiling of the mitochondrial proteome using affinity fractionation and automation. Electrophoresis 21:3427–3440
DiMauro S, Schon EA (2003) Mitochondrial respiratory-chain diseases. N Engl J Med 348:2656–2668
Milone M, Benarroch EE (2012) Mitochondrial dynamics: general concepts and clinical implications. Neurology 78:1612–1619
Ajroud-Driss S, Fecto F, Ajroud K, Siddique T (2012) Mutations in the nuclear encoded novel mitochondrial protein CHCHD10 cause an autosomal dominant mitochondrial myopathy. Neurology 78
Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, Hurles ME, McVean GA (2010) A map of human genome variation from population-scale sequencing. Nature 467:1061–1073
Heiman-Patterson TD, Argov Z, Chavin JM, Kalman B, Alder H, DiMauro S, Bank W, Tahmoush AJ (1997) Biochemical and genetic studies in a family with mitochondrial myopathy. Muscle Nerve 20:1219–1224
Dagda RK, Cherra SJ 3rd, Kulich SM, Tandon A, Park D, Chu CT (2009) Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem 284:13843–13855
Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H et al (2011) Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477:211–215
Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK et al (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134:112–123
Claros MG, Vincens P (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:779–786
Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR et al (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39:D225–229
Baughman JM, Nilsson R, Gohil VM, Arlow DH, Gauhar Z, Mootha VK (2009) A computational screen for regulators of oxidative phosphorylation implicates SLIRP in mitochondrial RNA homeostasis. PLoS Genet 5:e1000590
Martherus RS, Sluiter W, Timmer ED, VanHerle SJ, Smeets HJ, Ayoubi TA (2010) Functional annotation of heart enriched mitochondrial genes GBAS and CHCHD10 through guilt by association. Biochem Biophys Res Commun 402:203–208
Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ Jr (2005) Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol 6:R33
Roth RB, Hevezi P, Lee J, Willhite D, Lechner SM, Foster AC, Zlotnik A (2006) Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics 7:67–80
Darshi M, Mendiola VL, Mackey MR, Murphy AN, Koller A, Perkins GA, Ellisman MH, Taylor SS (2011) ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J Biol Chem 286:2918–2932
Milone M, Wong LJ (2013) Diagnosis of mitochondrial myopathies. Mol Genet Metab 110:35–41
Bannwarth S, Ait-El-Mkadem S, Chaussenot A, Genin EC, Lacas-Gervais S, Fragaki K, Berg-Alonso L, Kageyama Y, Serre V, Moore DG et al (2014) A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain 137:2329–2345
Herrmann JM, Kohl R (2007) Catch me if you can! Oxidative protein trapping in the intermembrane space of mitochondria. J Cell Biol 176:559–563
Longen S, Bien M, Bihlmaier K, Kloeppel C, Kauff F, Hammermeister M, Westermann B, Herrmann JM, Riemer J (2009) Systematic analysis of the twin cx(9)c protein family. J Mol Biol 393:356–368
Banci L, Bertini I, Ciofi-Baffoni S, Tokatlidis K (2009) The coiled coil-helix-coiled coil-helix proteins may be redox proteins. FEBS Lett 583:1699–1702
Cavallaro G (2010) Genome-wide analysis of eukaryotic twin CX9C proteins. Mol BioSyst 6:2459–2470
An J, Shi J, He Q, Lui K, Liu Y, Huang Y, Sheikh MS (2012) CHCM1/CHCHD6, novel mitochondrial protein linked to regulation of mitofilin and mitochondrial cristae morphology. J Biol Chem 287:7411–7426
Acknowledgments
We thank the patients and their family members for participating in this study. We also thank Kreshnik B. Ahmeti, LiJun Cheng, and Yi Yang for their technical assistance. This work was supported by the National Institute of Neurological Disorders and Stroke (NS050641); the Les Turner ALS Foundation/Herbert C. Wenske Foundation Professorship; the David C. Asselin MD Memorial Fund; the Vena E. Schaff ALS Research Fund; the George Link, Jr. Foundation; and the Foglia Family Foundation. This work was supported by a grant (R01GM097136) from the National Institutes of Health to VKM. Imaging work was performed at the Northwestern University Cell Imaging Facility generously supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center.
Conflict of interest
None
Author information
Authors and Affiliations
Corresponding authors
Additional information
Senda Ajroud-Driss and Faisal Fecto contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
(DOCX 3591 kb)
Supplementary Figure 2
(DOCX 229 kb)
Supplementary Figure 3
(DOCX 78 kb)
Supplementary Figure 4
(DOCX 349 kb)
Supplementary Table 1
(DOCX 21 kb)
Supplementary Table 2
(DOCX 18 kb)
Rights and permissions
About this article
Cite this article
Ajroud-Driss, S., Fecto, F., Ajroud, K. et al. Mutation in the novel nuclear-encoded mitochondrial protein CHCHD10 in a family with autosomal dominant mitochondrial myopathy. Neurogenetics 16, 1–9 (2015). https://doi.org/10.1007/s10048-014-0421-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-014-0421-1