Abstract
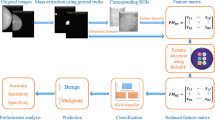
Histopathology is the gold standard for accurate diagnosis of cancer, tumors and similar diseases. Real-world pathological images, due to non-homogeneous nature and unorganized spatial intensity variations, are complex to analyze and classify. The major challenge in classifying pathological images is the complexity due to high intra-class variability and low inter-class variation in texture. Accuracy of histopathological image classification is highly dependent on the relevancy of the selected features to the problem. This paper is an effort in the same direction and presents an abstract feature based framework called abstract feature framework (AFF) to select optimal set of the most relevant features to classify pathological images. An abstract feature is created by identifying interlinked run-length texture features and grouping them. AFF is comprised of a new data structure called Abstract Feature Tree (AFT) and an algorithm for manipulating it. AFT is a tree structure in which nodes are abstract features. The Linkage Learning Algorithm for manipulating AFT is the brain of this framework and inspired by genetic algorithm. It creates better abstract features by first identifying interlinked abstract features and then combining them. This process is repeated until no improvement is found. On termination, the final list of abstract features is used for classifying pathological images. The proposed framework was tested on real-world histopathological meningioma dataset. Results obtained proved that the proposed framework outperformed the best-known rank-based feature selection techniques by using, on average, approximately three times less features to achieve 22% higher classification accuracy.











Similar content being viewed by others
References
Al-Janabi S, Huisman A, Van Diest PJ (2012) Digital pathology: current status and future perspectives. Histopathology 61(1):1–9
Al-Kadi O (2010) Texture measures combination for improved meningioma classification of histopathological images. Pattern Recogn 43(6):2043–2053
Al-Kadi OS (2014) A multiresolution clinical decision support system based on fractal model design for classification of histological brain tumours. Comput Med Imaging Graph 41:67–79
Behroozmand R, Almasganj F (2007) Optimal selection of wavelet-packet-based features using genetic algorithm in pathological assessment of patients’ speech signal with unilateral vocal fold paralysis. Comput Biol Med 37(4):474–485
Bloom F, Beal F, Kupfer D (2002) The Dana guide to brain health, 1st edn. Simon and Schuster Dana Press, New York
Boser B, Guyon I, Vapnik V (1992) A training algorithm for optimal margin classifiers. In: Proceedings of the fifth annual workshop on Computational learning theory, pp 144–152. ACM
Breiman L (2001) Random forests. Mach Learn 45(1):5–32
Chekkoury A, Khurd P, Ni J, Bahlmann C, Kamen A, Patel A, Grady L, Singh M, Groher M, Navab N, et al (2012) Automated malignancy detection in breast histopathological images. In: SPIE Medical Imaging, pp 831,515–831,515. International Society for Optics and Photonics
Cruz-Roa A, Caicedo JC, González FA (2011) Visual pattern mining in histology image collections using bag of features. Artif Intell Med 52(2):91–106
Deepa S, Devi BA (2011) A survey on artificial intelligence approaches for medical image classification. Indian J Sci Technol 4(11):1583–1595
Dong F, Irshad H, Oh EY, Lerwill MF, Brachtel EF, Jones NC, Knoblauch NW, Montaser-Kouhsari L, Johnson NB, Rao LKF, Faulkner-Jones B, Wilbur DC, Schnitt SJ, Beck AH (2014) Computational pathology to discriminate benign from malignant intraductal proliferations of the breast. PLoS ONE 9(12):e114,885. doi:10.1371/journal.pone.0114885
Dundar MM, Badve S, Bilgin G, Raykar V, Jain R, Sertel O, Gurcan MN (2011) Computerized classification of intraductal breast lesions using histopathological images. IEEE Trans Biomed Eng 58(7):1977–1984
Fatima K, Arooj A, Majeed H (2014) A new texture and shape based technique for improving meningioma classification. Microsc Res Tech 77(11):862–873
Galar M, Fernández A, Barrenechea E, Bustince H, Herrera F (2011) An overview of ensemble methods for binary classifiers in multi-class problems: experimental study on one-vs-one and one-vs-all schemes. Pattern Recogn 44(8):1761–1776
Gonzalez RC, Richard E (2002) Woods, digital image processing. Prentice Hall Press, Englewood Cliffs. ISBN: 0-201-18075-8
Gorelick L, Veksler O, Gaed M, Gomez J, Moussa M, Bauman G, Fenster A, Ward A (2013) Prostate histopathology: Learning tissue component histograms for cancer detection and classification. IEEE Trans Med Imaging 32(10):1804–1818
Gurcan M, Boucheron L, Can A, Madabhushi A, Rajpoot N, Yener B (2009) Histopathological image analysis: a review. IEEE Rev Biomed Eng 2:147–171
Hall MA (1999) Correlation-based feature selection for machine learning. Technical report. The University of Waikato, Hamilton
Hsu CW, Lin CJ (2002) A comparison of methods for multiclass support vector machines. IEEE Trans Neural Netw 13(2):415–425
Huang PW, Lai YH (2010) Effective segmentation and classification for HCC biopsy images. Pattern Recogn 43(4):1550–1563
Irshad H (2013) Automated mitosis detection in histopathology using morphological and multi-channel statistics features. J Pathol Inf 4(1):10
Irshad H, Gouaillard A, Roux L, Racoceanu D (2014) Multispectral band selection and spatial characterization: application to mitosis detection in breast cancer histopathology. Comput Med Imaging Graph 38(5):390–402
Irshad H, Veillard A, Roux L, Racoceanu D (2014) Methods for nuclei detection, segmentation, and classification in digital histopathology: a review, current status and future potential. IEEE Rev Biomed Eng 7:97–114. doi:10.1109/RBME.2013.2295804
Jung S, Qiao X (2014) A statistical approach to set classification by feature selection with applications to classification of histopathology images. Biometrics 70(3):536–545
La Cava W, Danai K, Spector L, Fleming P, Wright A, Lackner M (2016) Automatic identification of wind turbine models using evolutionary multiobjective optimization. Renew Energy 87:892–902
Lauro GR, Cable W, Lesniak A, Tseytlin E, McHugh J, Parwani A, Pantanowitz L (2013) Digital pathology consultations—a new era in digital imaging, challenges and practical applications. J Digit Imaging 26(4):668–677
Lee J (2009) Meningiomas: diagnosis, treatment, and outcome. Springer, Berlin
Lee JH (2008) Meningiomas: diagnosis, treatment, and outcome. Springer, Berlin
Liu H, Setiono R, et al (1996) A probabilistic approach to feature selection—a filter solution. In: ICML, vol 96, pp 319–327. Citeseer
Loménie N, Racoceanu D (2012) Point set morphological filtering and semantic spatial configuration modeling: application to microscopic image and bio-structure analysis. Pattern Recogn 45(8):2894–2911
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P (2007) The 2007 who classification of tumours of the central nervous system. Acta Neuropathol 114(2):97–109
Mallat SG (1989) A theory for multiresolution signal decomposition: the wavelet representation. IEEE Trans Pattern Anal Mach Intell 11(7):674–693
Maulik U (2009) Medical image segmentation using genetic algorithms. IEEE Trans Inf Technol Biomed 13(2):166–173
Milner S, Davis C, Zhang H, Llorca J (2012) Nature-inspired self-organization, control, and optimization in heterogeneous wireless networks. IEEE Trans Mob Comput 11(7):1207–1222
Peng H, Long F, Ding C (2005) Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell 27(8):1226–1238
Qureshi H, Rajpoot N, Nattkemper T, Hans V (2009) A robust adaptive wavelet-based method for classification of meningioma histology images. In: Proceedings MICCAI’2009 Workshop on Optical Tissue Image Analysis in Microscopy, Histology, and Endoscopy (OPTIMHisE)
Serra J (1982) Image analysis and mathematical morphology. Academic Press, London [Review by Fensen EB in J Microsc 131:258 (1983)] Cell size, Staining Microscopy Technique, Mathematics, General article Review article (PMBD 185707888)
Siddiqi K, Shokoufandeh A, Dickinson SJ, Zucker SW (1999) Shock graphs and shape matching. Int J Comput Vision 35(1):13–32
Sternberg SR (1986) Grayscale morphology. Comput Vision Graph Image Process 35(3):333–355
Tang X (1998) Texture information in run-length matrices. IEEE Trans Image Process 7(11):1602–1609
Tax DM, Duin RP (2002) Using two-class classifiers for multiclass classification. In: Pattern Recognition, 2002. Proceedings. 16th International Conference on, vol 2, pp 124–127. IEEE
Vapnik V (1999) The nature of statistical learning theory. Springer, Berlin
Wiemels J, Wrensch M, Claus EB (2010) Epidemiology and etiology of meningioma. J Neurooncol 99(3):307–314
Yagi Y (2011) Color standardization and optimization in whole slide imaging. Diagn Pathol 6(Suppl 1):S15
Acknowledgements
The authors would like to thank Dr. Nasir M. Rajpoot, Associate Professor, Department of Computer Science, University of Warwick, the UK, for the provision of meningioma dataset of the Institute of Neuropathology, Bielefeld, Germany.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Majeed, H., Fatima, K. Synergies between texture features: an abstract feature based framework for meningioma subtypes classification. Pattern Anal Applic 20, 1209–1225 (2017). https://doi.org/10.1007/s10044-017-0599-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10044-017-0599-6