Abstract
In this paper, we propose a brain tumor segmentation and classification method for multi-modality magnetic resonance imaging scans. The data from multi-modal brain tumor segmentation challenge (MICCAI BraTS 2013) are utilized which are co-registered and skull-stripped, and the histogram matching is performed with a reference volume of high contrast. From the preprocessed images, the following features are then extracted: intensity, intensity differences, local neighborhood and wavelet texture. The integrated features are subsequently provided to the random forest classifier to predict five classes: background, necrosis, edema, enhancing tumor and non-enhancing tumor, and then these class labels are used to hierarchically compute three different regions (complete tumor, active tumor and enhancing tumor). We performed a leave-one-out cross-validation and achieved 88% Dice overlap for the complete tumor region, 75% for the core tumor region and 95% for enhancing tumor region, which is higher than the Dice overlap reported from MICCAI BraTS challenge.
Similar content being viewed by others
1 Introduction
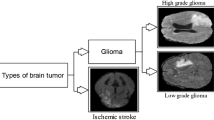
The detection and diagnosis of brain tumor from MRI is crucial to decrease the rate of casualties. Brain tumor is difficult to cure, because the brain has a very complex structure and the tissues are interconnected with each other in a complicated manner. Despite many existing approaches, robust and efficient segmentation of brain tumor is still an important and challenging task. Tumor segmentation and classification is a challenging task, because tumors vary in shape, appearance and location. It is hard to fully segment and classify brain tumor from mono-modality scans, because of its complicated structure. MRI provides the ability to capture multiple images known as multi-modality images, which can provide the detailed structure of brain to efficiently classify the brain tumor [1]. Figure 1 shows different MRI modalities of brain.
Brain tumor segmentation and detailed classification based on MRI images has received considerable interest over last decades. It has been explored in many studies using uni-modality MRI. Recently, researchers have explored multi-modality MRI to increase the accuracy of tumor segmentation and classification.
Machine learning and edge/region-based approaches have been used with multi-modality (T1, T2, T1C and FLAIR) MRI [2]. The machine learning techniques often rely on voxel intensities and texture features. Individual voxel is classified on the basis of feature vector [2]. Intensity, intensity difference, neighborhood and other texture features have been explored on benchmark dataset [3]. To the best of our knowledge, wavelet-based features have not yet been explored on multi-modality MRI brain tumor dataset. In this paper, we investigate wavelet texture features along with various machine learning algorithms.
In this work, we used multi-modality images to classify the brain tumor. This work makes the following contributions:
-
1.
extracting wavelet-based texture features to predict tumor labels and
-
2.
exploring supervised classifiers for brain tumor classification.
This paper is organized as follows: Sect. 2 reviews the related work; Sect. 3 discusses the proposed algorithm, while Sect. 4 presents the results, leading to conclusion in Sect. 5.
2 Literature review
Brain tumor segmentation is a challenging process because tumor exhibits inhomogeneous intensities and unclear boundaries. Intensity normalization or bias field correction is often applied to balance the effect of magnetic field inhomogeneity [1]. Intensities, neighborhood and texture are common features used in various studies [1,2,3]. Various machine learning and edge/region-based techniques used in segmentation are summarized in Table 1, where we present a concise review of the previous work. Few techniques are fully automatic, while remaining need user involvement.
Fluid vector flow (FVF) [4] is introduced to address the problem of unsatisfactory capture range and poor convergence for concavities. Harati et al. [5] demonstrated an improved fuzzy connectedness (FC) algorithm, where seed points are selected automatically to segment the tumor region. Saha et al. [6] proposed a fast novel method to locate the bounding box around tumor or edema using Bhattacharya coefficient [7]. In their proposed clustering technique axial view of brain image is divided into left and right halves, and then a rectangle is used to compare the corresponding regions of left half with right half to find the most dissimilar region within the rectangle. Zhu et al. [8] proposed a semiautomatic brain tumor segmentation method, where initial segmentation is performed through ITK-Snap tool. Voxel-based segmentation and deformable shape-based segmentation are combined into the software pipeline. Sachdeva et al. [9] used texture information with intensity in active contour model (ACM) to overcome the issue observed in previous techniques like FVF, boundary vector flow (BVF) and gradient vector flow (GVF). In previous techniques selection of false edges or false seeds corresponds to preconvergence problem and selection of weak edges leads to over-segmentation due to the edema around the tumor. Rexilius et al. [10] proposed a new region growing method for segmentation of brain tumor. Probabilistic model is used to achieve the initial segmentation, which is further refined by region growing to give better segmentation results. Global affine and non-rigid registration method is used to register multi-spectral histograms gathered from patients’ data with a reference histogram.
Corso et al. [11] used a top-down approach to distribute the product over generative model. Later, sparse graph is given as input to graph cut method, where each edge uses features to find similarity between neighboring nodes having the affinity. Segmentation by weighted aggregation (SWA) is used to provide the multi-level segmentation of data. Ruan et al. [12] proposed a supervised machine learning technique to track the tumor volume. The complete process is categorized into two main steps. In the first step to make it efficient and reduce computational time, only T1 modality is used to identify the abnormal area. In the second step, the abnormal area is extracted from all modalities and fused to segment the tumor. Irfan et al. [13] introduced a technique in which brain images are separated from non-brain part, and then ROI is used with the saliency information to bind the search of normalization cut (N-Cut) [14] method. Saliency information is the combination of multi-scale contrast and image curvature points. Multi-scale contrast image is acknowledged when image is decomposed at multiple scales by using Gaussian pyramid (GP), and Euclidean distance is calculated with neighboring pixels at those scales.
Automatic segmentation is performed using the random forest (RF) [3], where features include MR sequence intensities, neighborhood information, context information and texture. Post-processing is performed for the sake of good results. Zhao et al. [15] used Markov random field (MRF) model on supervoxels to automatically segment tumor. ACM combines the edge-based and region-based techniques [16], where user draws region of interest (ROI) in different images on the basis of tumor type and grade.
In machine learning availability of benchmark data became important in comparing different algorithms. Recently, this idea has also become popular in the domain of medical image analysis. Sometime challenge word is used instead of benchmark that shares the common characteristic in a sense that different researchers used their own algorithms to optimize on a training dataset provided by the organizers of event and then apply their algorithm to a common, independent test dataset. The benchmark idea is different from other published comparisons in a sense that in benchmark each group of researchers uses the same dataset for their algorithm. The BraTS benchmark was established in 2012, and first event was held in the same year [2]. Dataset consists of real and simulated images.
Various studies presented different accuracy measures and dataset as shown in Table 1; therefore, it is difficult to compare them and draw conclusion about the best technique. Furthermore, in previous studies, the value of Dice and Jaccard was not high enough and there is room for further improvement in classification accuracy; therefore, we explored wavelet-based texture features which were not explored before on MICCAI BraTS dataset.
3 Proposed method
The proposed algorithm uses MICCAI BraTS dataset and the main flow of our proposed technique is presented in Fig. 2, with further details presented in subsection.
3.1 Preprocessing
The BraTS dataset has four modalities of MRI: T1, T2, T1C and FLAIR. Each modality scan is rigidly co-registered with T1C modality to homogenize data, because T1C has the highest spatial resolution in most cases. Linear interpolator is used to resample all the images to 1-mm isotropic resolution in axial orientation. Images are skull-stripped with expert annotation [2]. All the images are visualized through ITK-Snap [17], while histogram matching is performed with Slicer3D [18] to enhance the image contrast by choosing a high-contrast image as the reference.
The next preprocessing step is to determine the bounding box around the tumor region. Our adapted technique for locating bounding box consists of the following steps:
-
1.
Remove complete blank slices from ground truth, remaining slices contain tumor part.
-
2.
Create a mask and use it to locate bounding box in ground truth.
-
3.
Use the above bounding box to crop multi-modality images.
3.2 Feature extraction
The proposed feature extraction includes four types of features: (1) intensity, (2) intensity difference, (3) neighborhood information and (4) wavelet-based texture features.
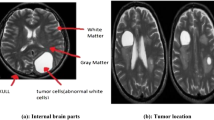
Intensity features are shown Fig. 1. Intensity difference is the differences between the above modalities, and we used three prominent intensity difference features that represent the global characteristics of brain tissues [19] as shown in Fig. 3.
Neighborhood information features include mean, median and range of 3D neighbors centered at voxel being considered. The isotropic neighborhood size of 3, 9, 15 and 19 mm was used in 3D as these were found to be appropriate for mean and range filters [3], while we used median filter with neighborhood size 3 mm.
The novelty of the proposed approach is to extract wavelet features, which has not been explored and applied on MICCAI BraTS dataset. Wavelet has the property of multi-resolution analysis, where we can decompose and visualize the images at different scales [20]. Discrete wavelet transform can be defined as:
where \(j,k \in Z\), j controls the dilation, k controls the translation of wavelet function, and \(\vartheta \left( t \right)\) is the mother wavelet. Performing scaling and shifting on initial wavelet and convolving it with the original image is a part of wavelet decomposition. It has the property to reconstruct the original image without loss of information [21]. Wavelet-based texture segmentation is compared with simple single resolution texture spectrum, co-occurrences and local linear transforms on Brodatz dataset, where wavelet-based texture segmentation performed better than other approaches [22]. Wavelet has been used on brain, liver and kidney 3D images to produce accurate reconstruction from decomposed subimages [23].
For 3D wavelet decomposition, the image volume is initially convolved in x dimension with low-pass filter to produce approximation subband (L) and with high-pass filter to produce detail subband (H). In the same way, the approximation and detail subbands are further convolved in y dimension and z dimension, respectively, with both the low-pass and high-pass filters. As a result, eight subbands: LLL, LLH, LHL, HLL, LHH, HLH, HHL and HHH [21] are obtained, where L indicates low-pass-filtered subband and H indicates high-pass-filtered subband. Level 2 decomposition is achieved by considering the LLL subband as the main image and decomposing with the same process as above.
Block diagram of wavelet-based feature extraction is shown in Fig. 4. In wavelet-based feature extraction, an intensity difference image (from T1C, T1C-FLAIR, T1C-T1 or T2-T1C) is given as input for 3D wavelet decomposition. Input image is decomposed into subbands, and subbands containing useful information are then selected based on their discriminatory ability assessed by visual analysis. Feature images are reconstructed from selected subband, and Gaussian filter is applied after absolute function to make the features more prominent. We performed decomposition at second level, because subbands of third level were not found to be useful in our experiments. Moreover, the subbands at third level of decomposition are at too small scale to contain sufficiently useful discriminatory information. We tried various filter families for wavelet decomposition including Daubechies4, Symlets4 and Symlets8, while Symlets8 was selected due to superior performance.
Block diagram of wavelet-based feature extraction, while input to wavelet decomposition can be intensity differences or T1C modality and output represents the feature images [24]
Wavelet reconstruction is a process in which feature images are constructed from each subband, and useful feature images are then selected based on discriminatory information present in visual analysis. We applied absolute function and Gaussian smoothing to make the edges of feature images more prominent [24] as shown in Fig. 5.
In this work, we extracted intensity, intensity differences, neighborhood information and wavelet-based texture features. In the next section, we will use these features to perform supervised classification.
3.3 Classification
Supervised classification is a machine learning approach in which training data are used to construct the model and test data are used to evaluate the constructed model on unseen data to measure the performance of algorithm. There are a number of classifiers that exist to classify data, and below we will discuss the classifiers which we have explored in this work.
The kNN (k-nearest neighbor) is a lazy learning technique, which calculates the Euclidean distance from all the points. The classification label is then assigned based upon majority voting as per ‘k’ nearest neighbors.
Random forest (RF) is a combination of decision trees. Each tree in ensemble is trained on randomly sampled data with replacement from training vector during the phase of training. Multiple trees are trained to increase the correlation and reduce the variance between trees. In test phase, vote of each tree is considered and majority vote is given to the unseen data. RF is useful because it gives internal estimates of error and variable importance, and also it can be easily parallelized [25]. RF has become a major data analysis tool within a short period of time, and it became popular because it can be applied to nonlinear and higher-order dataset [26].
AdaBoostM2 (adaptive boosting) [27] is the enhanced version of AdaBoostM1 [27], which is used for multi-class classification. It is a boosting algorithm, where many weak learners are combined to make a powerful algorithm and instances are reweighted rather than resampled (in bagging) [25].
Random under sampling (RusBoost) is suitable for classifying imbalanced data when instances of one class dominate many times than the other. Machine learning techniques fail to efficiently classify skewed data, but RusBoost solved the problem by combining sampling and boosting. We explored these classification algorithms, and the results are reported in the next section.
4 Results
In this section, we present the results and compare them with previous work on the BraTS dataset of real patients containing 20 high-grade (HG) and 10 low-grade (LG) subjects. Three measures are used for quantitative evaluation, and visual segmentation results are also shown. The results are obtained on HP-probook 4540, Core i5, 2.5 GHz, 8 GB RAM using MATLAB 2013a, and it takes about 2 min to test a new patient.
4.1 Out of bag error (ooBError)
OoBError is the mean-squared error or the misclassification error for out of bag observations in the training. There is no need of separate test set of cross-validation to get the unbiased estimated error for test cases, because ooBError is calculated internally during RF model creation phase. Figure 6 shows that ooBError is lowest when 25 trees are used.
4.2 Evaluation measures
We used various evaluation measures to assess the results, and these measures are described below. The Dice coefficient is the similarity/overlap between two images [28]. It is graphically explained in Fig. 7:
where \(\cap\) is the logical AND operator, | | is the size of the set (i.e., the number of voxels belonging to it). \(P_{1}\) and \(T_{1}\) represent the numbers of voxels belonging to algorithm’s prediction and ground truth, respectively. The Dice score normalizes the number of true positives to the average size of predicted and ground truth-segmented area. It also gives us the voxel wise overlap between the result and ground truth [2].
Dice score is calculated by deriving formula from the diagram. \(T_{1}\) is the ground truth lesion, and \(T_{0}\) is the area outside \(T_{1}\) within the brain. \(P_{1}\) is the algorithm’s predicted lesion, and \(P_{0}\) is the algorithm’s predicted area outside \(P_{1}\) within the brain. Overlapped area between \(T_{1}\) and \(P_{1}\) gives us the true positive [2]
The Jaccard coefficient measures the similarity between two images and can be defined as the size of intersection divided by the size of union of two sets [29]. Jaccard coefficient is also known as Jaccard index and can be measured as:
Sensitivity is true positive rate, it is prioritized when disease is serious, and we want to identify all the possible true cases. It can be measured as:
Specificity is true negative rate, it is prioritized when treatment is dreadful, and we only want to treat those which are surely having disease. It can be measured:
4.3 Hierarchical classification
Each voxel is initially classified as one of the five target classes [background (0), necrosis (1), edema (2), non-enhancing (3) and enhancing (4)]. Subsequently, tumor regions are computed hierarchically from these class labels. Our classification system extracts the following three tumor regions in a hierarchical manner:
-
1.
Complete Tumor: This region is the combination of four classes (1) + (2) + (3) + (4), which are separated from class (0).
-
2.
Core Tumor: In this region, we exclude edema (2) from complete tumor identified in step above.
-
3.
Enhancing Tumor: Subsequent to core tumor classification, enhancing tumor (4) is extracted from necrosis and non-enhancing (1) + (3).
For our initial experiments, in order to identify experimental choices, we performed leave-one-out cross-validation on a subset of BraTS data (four real HG patients) with the assumption that the identified choices will perform similar on complete BraTS data. The initial experiments on a subset of data were conducted for computational reasons. Table 2 presents the comparison between different types of features and shows that wavelet features are helpful in improving Dice coefficient. We utilized all the extracted features to compare different classifiers as shown in Table 3.
4.4 Quantitative evaluation
Table 3 shows that RF is performing best among other classifiers for the extracted features, therefore we used RF classifier, and the quantitative results of the proposed method are compared with the results presented by the MICCAI BraTS challenge in Table 4. Table 5 shows the detail results of proposed methodology.
4.5 Visual results
Visual results of the work are shown in Fig. 8, indicating the success of brain tumor classification with the proposed method.
5 Discussion
We proposed an algorithm for brain tumor classification. The proposed algorithm used MICCAI BraTS data and relies on intensity-related features and wavelet texture features. The algorithm is applied on BraTS challenge training dataset, and it gives better results than the state-of-the-art methods as shown in Table 4.
In feature extraction process, we calculated intensity, intensity difference and neighborhood information features [3] and the wavelet texture features. For wavelet features, we initially decomposed the multi-modality images into third level and visualized all the feature images produced by these. We restrict wavelet decomposition at second level after visualization, because the feature images at third level are too small and not much useful for us. We analyzed all the feature images at first and second level and selected only those, which contain high-frequency components. Future work will focus on improving subband selection process to make it more automatic rather than based on visualization and to test the algorithm on larger dataset to verify robustness.
We utilized all the extracted features with different classifiers (kNN, RF, AdaBoostM2 and RusBoost) as in Table 3 and observed that RF is better for our extracted features to classify brain tumor. Leave-one-out cross-validation is performed separately for HG and LG on real dataset. We further performed detailed classification that classifies the tumor into three different regions: complete tumor, core tumor and enhancing tumor. Proposed technique gives comparable or favorable results with other existing techniques.
6 Conclusion
This paper presented an algorithm to hierarchically classify the tumor into three regions: whole tumor, core tumor and enhancing tumor. Intensity, intensity difference, neighborhood information and wavelet features are extracted and utilized on multi-modality MRI scans with various classifiers. The use of wavelet-based texture features with RF classifier has increased the classification accuracy as evident by quantitative results of our proposed method which are comparable or higher than the state of the art.
References
Bauer S, Wiest R, Nolte L-P, Reyes M (2013) A survey of MRI-based medical image analysis for brain tumor studies. Phys Med Biol 58:R97
Menze BH, Jakab A, Bauer S et al (2015) The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging 34(10):1993–2024
Festa J, Pereira S, Mariz JA, Sousa N, Silva CA (2013) Automatic brain tumor segmentation of multi-sequence mr images using random decision forests. In Proceedings MICCAI BRATS, 2013
Wang T, Cheng I, Basu A (2009) Fluid vector flow and applications in brain tumor segmentation. IEEE Trans Biomed Eng 56:781–789
Harati V, Khayati R, Farzan A (2011) Fully automated tumor segmentation based on improved fuzzy connectedness algorithm in brain MR images. Comput Biol Med 41:483–492
Saha BN, Ray N, Greiner R, Murtha A, Zhang H (2012) Quick detection of brain tumors and edemas: a bounding box method using symmetry. Comput Med Imaging Graph 36:95–107
Khalid MS, Ilyas MU, Sarfaraz MS, Ajaz MA (2006) Bhattacharyya coefficient in correlation of gray-scale objects. J Multimed 1:56–61
Zhu Y, Young GS, Xue Z, Huang RY, You H, Setayesh K, Hatabu H, Cao F, Wong ST (2012) Semi-automatic segmentation software for quantitative clinical brain glioblastoma evaluation. Acad Radiol 19:977–985
Sachdeva J, Kumar V, Gupta I, Khandelwal N, Ahuja CK (2012) A novel content-based active contour model for brain tumor segmentation. Magn Reson Imaging 30:694–715
Rexilius J, Hahn HK, Klein J, Lentschig MG, Peitgen H-O (2007) Multispectral brain tumor segmentation based on histogram model adaptation. In: Medical imaging—SPIE, 2007, pp 65140V–65140V-10
Corso JJ, Sharon E, Dube S, El-Saden S, Sinha U, Yuille A (2008) Efficient multilevel brain tumor segmentation with integrated bayesian model classification. IEEE Trans Med Imaging 27:629–640
Ruan S, Lebonvallet S, Merabet A, Constans J (2007) Tumor segmentation from a multispectral MRI images by using support vector machine classification. In: 4th IEEE international symposium on Biomedical imaging: from nano to macro, 2007. ISBI 2007, pp 1236–1239
Mehmood I, Ejaz N, Sajjad M, Baik SW (2013) Prioritization of brain MRI volumes using medical image perception model and tumor region segmentation. Comput Biol Med 43:1471–1483
Shi J, Malik J (2000) Normalized cuts and image segmentation. IEEE Trans Pattern Anal Mach Intell 22:888–905
Zhao L, Sarikaya D, Corso JJ (2013) Automatic brain tumor segmentation with MRF on supervoxels. In: Proceedings of NCI-MICCAI BRATS, vol 1, p 51
Guo X, Schwartz L, Zhao B (2013) Semi-automatic segmentation of multimodal brain tumor using active contours. In: Proceedings MICCAI BRATS, 2013
Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31:1116–1128
Kikinis R, Pieper S (2011) 3D Slicer as a tool for interactive brain tumor segmentation. In: Engineering in medicine and biology society, EMBC, 2011 annual international conference of the IEEE, 2011, pp 6982–6984
Reza S, Iftekharuddin K (2013) Multi-class abnormal brain tissue segmentation using texture features. In: Proceedings of NCI-MICCAI BRATS, vol 1, pp 38–42
Sifuzzaman M, Islam M, Ali M (2009) Application of wavelet transform and its advantages compared to Fourier transform. J Phys Sci 13:121–134
Procházka A, Gráfová L, Vyšata O, Caregroup N (2011) Three-dimensional wavelet transform in multi-dimensional biomedical volume processing. In: Proceedings of the IASTED international conference graphics and virtual reality, Cambridge, UK, 2011
Arivazhagan S, Ganesan L (2003) Texture segmentation using wavelet transform. Pattern Recogn Lett 24:3197–3203
Cheng J, Liu Y (2009) 3-D reconstruction of medical image using wavelet transform and snake model. J Multimed 4:427–434
Rajpoot KM, Rajpoot NM (2004) Wavelets and support vector machines for texture classification. In Multitopic conference, 2004. Proceedings of INMIC 2004. 8th international, 2004, pp 328–333
Breiman L (2001) Random forests. Mach Learn 45:5–32
Strobl C, Boulesteix A-L, Zeileis A, Hothorn T (2007) Bias in random forest variable importance measures: illustrations, sources and a solution. BMC Bioinform 8:25
Freund Y, Schapire RE (1997) A decision-theoretic generalization of on-line learning and an application to boosting. J Comput Syst Sci 55:119–139
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302
Jaccard P (1912) The distribution of the flora in the alpine zone. New Phytol 11:37–50
Bauer S, Nolte L-P, Reyes M (2011) Fully automatic segmentation of brain tumor images using support vector machine classification in combination with hierarchical conditional random field regularization. In: Medical image computing and computer-assisted intervention–MICCAI 2011. Springer, Lecture Notes in Computer Science, vol 6893, pp 354–361
Doyle S, Vasseur F, Dojat M, Forbes F (2013) Fully automatic brain tumor segmentation from multiple MR sequences using hidden Markov fields and variational EM. In: Proceedings of the NCI-MICCAI BraTS, pp 18–22
Cordier N, Menze B, Delingette H, Ayache N (2013) Patch-based segmentation of brain tissues. In MICCAI challenge on multimodal brain tumor segmentation, 2013, pp 6–17
Subbanna NK, Precup D, Collins DL, Arbel T (2013) Hierarchical probabilistic gabor and MRF segmentation of brain tumours in MRI volumes. In: Medical image computing and computer-assisted intervention–MICCAI 2013. Springer, Lecture Notes in Computer Science, vol 8149, pp 751–758
Tustison N, Wintermark M, Durst C, Avants B (2013) ANTs and arboles. In: Proceedings of NCI-MICCAI BRATS, vol 1, p 47
Wu W, Chen AY, Zhao L, Corso JJ (2014) Brain tumor detection and segmentation in a CRF (conditional random fields) framework with pixel-pairwise affinity and superpixel-level features. Int J Comput Assist Radiol Surg 9:241–253
Acknowledgements
We would like to thank the organizers of MICCAI BraTS 2013 challenge for sharing the dataset. Brain tumor image data used in this work were obtained from the NCI-MICCAI 2013 Challenge on Multimodal Brain Tumor Segmentation (http://martinos.org/qtim/miccai2013/index.html) organized by K. Farahani, M. Reyes, B. Menze, E. Gerstner, J. Kirby and J. Kalpathy-Cramer. The challenge database contains fully anonymized images from the following institutions: ETH Zurich, University of Bern, University of Debrecen and University of Utah and publicly available images from the Cancer Imaging Archive (TCIA).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Usman, K., Rajpoot, K. Brain tumor classification from multi-modality MRI using wavelets and machine learning. Pattern Anal Applic 20, 871–881 (2017). https://doi.org/10.1007/s10044-017-0597-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10044-017-0597-8