Abstract
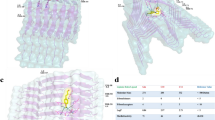
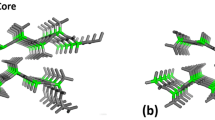
Understanding the nature of the recognition between amyloid protofibrils and dye molecules at the molecular level is essential to improving instructive guides for designing novel molecular probes or new inhibitors. However, the atomic details of the binding between dyes and amyloid fibrils are still not fully understood. In this study, molecular docking, consensus scoring, molecular dynamics (MD), and molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) analyses were integrated to investigate the binding between Congo red (CR) and the GNNQQNY protofibril from yeast prion protein Sup35 and to further evaluate their binding stabilities and affinities. Our results reveal that there are four CR binding sites located on GNNQQNY protofibril surface. These four CR binding sites adopt dual binding modes by which CR binding with its long axis parallel and perpendicular to the long axis of the protofibril. In addition, CR was also found to bind to the edge of the protofibril via hydrophobic/aromatic and hydrogen-bonding interactions, which is inferred as the possible inhibition mechanism to prevent the elongation of the protofibril from the addition of incoming peptides. Virtual screening from National Cancer Institute (NCI) database obtained three hit compounds with higher binding affinity than CR to the edge of the protofibril due to the fact that the central parts of these compounds are able to form additional hydrogen bonds with the protofibril. The results of the study could be useful for the development of new molecular probes or inhibitors for clinical applications.

Investigation of the Congo red binding toward GNNQQNY protofibril and in silico virtual screening for the identification of new aggregation inhibitors









Similar content being viewed by others
References
Pepys MB (2001) Pathogenesis, diagnosis and treatment of systemic amyloidosis. Philos Trans R Soc Lond Ser B 356:203–211
Dobson CM (2005) Prying into prions. Nature 435:747–749
Makin OS, Serpell LC (2005) X-ray diffraction studies of amyloid structure. Methods Mol Biol 299:67–80
Makin OS, Serpell LC (2005) Structures for amyloid fibrils. FEBS J 272:5950–5961
Nelson R, Sawaya MR, Balbirnie M, Madsen A, Riekel C, Grothe R, Eisenberg D (2005) Structure of the cross-β spine of amyloid-like fibrils. Nature 435:773–778
Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT (2007) Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 447:453–457
Esposito L, Pedone C, Vitagliano L (2006) Molecular dynamics analyses of cross-β-spine steric zipper models: β-sheet twisting and aggregation. Proc Natl Acad Sci USA 103:11533–11538
Howie AJ, Brewer DB (2009) Optical properties of amyloid stained by Congo red: history and mechanisms. Micron 40:285–301
Lorenzo A, Yankner BA (1994) β-amyloid neurotoxicity requires fibril formation and is inhibited by congo red. Proc Natl Acad Sci USA 91:12243–12247
Reinke AA, Gestwicki JE (2007) Structure-activity relationships of amyloid β-aggregation inhibitors based on curcumin: influence of linker length and flexibility. Chem Biol Drug Des 70:206–215
Raman EP, Takeda T, Klimov DK (2009) Molecular dynamics simulations of ibuprofen binding to Aβ Peptides. Biophys J 97:2070–2079
Chang WE, Takeda T, Raman EP, Klimov DK (2010) Molecular dynamics simulations of anti-aggregation effect of ibuprofen. Biophys J 98:2662–2670
Lemkul JA, Bevan DR (2010) Destabilizing Alzheimer’s Aβ42 protofibrils with Morin: mechanistic insights from molecular dynamics simulations. Biochemistry 49:3935–3946
Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, Vakser IA (1992) Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc Natl Acad Sci USA 89:2195–2199
Chen R, Weng Z (2003) A novel shape complementarity scoring function for protein-protein docking. Proteins 51:397–408
Wu G, Robertson DH, Brooks Iii CL, Vieth M (2003) Detailed analysis of grid-based molecular docking: a case study of CDOCKER─CHARMm-based MD docking algorithm. J Comput Chem 24:1549–1562
Krammer A, Kirchhoff PD, Jiang X, Venkatachalam CM, Waldman M (2005) LigScore: a novel scoring function for predicting binding affinities. J Mol Graph Model 23:395–407
Gehlhaar DK, Verkhivker GM, Rejto PA, Sherman CJ, Fogel DR, Fogel LJ, Freer ST (1995) Molecular recognition of the inhibitor AG-1343 by HIV-1 protease: conformationally flexible docking by evolutionary programming. Chem Biol 2:317–324
Jain AN (1996) Scoring noncovalent protein-ligand interactions: a continuous differentiable function tuned to compute binding affinities. J Comput Aided Mol Des 10:427–440
Muegge I, Martin YC (1999) A general and fast scoring function for protein-ligand interactions: a simplified potential approach. J Med Chem 42:791–804
Muegge I (2006) PMF scoring revisited. J Med Chem 49:5895–5902
Böhm HJ (1998) Prediction of binding constants of protein ligands: a fast method for the prioritization of hits obtained from de novo design or 3D database search programs. J Comput Aided Mol Des 12:309–323
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N log (N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Miyamoto S, Kollman PA (1992) SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem 13:952–962
Wang J, Morin P, Wang W, Kollman PA (2001) Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J Am Chem Soc 123:5221–5230
Rafi SB, Cui G, Song K, Cheng X, Tonge PJ, Simmerling C (2006) Insight through molecular mechanics Poisson-Boltzmann surface area calculations into the binding affinity of triclosan and three analogues for FabI, the E. coli enoyl reductase. J Med Chem 49:4574–4580
Nolde SB, Arseniev AS, Orekhov VY, Billeter M (2002) Essential domain motions in barnase revealed by MD simulations. Proteins 46:250–258
Zeng J, Li W, Zhao Y, Liu G, Tang Y, Jiang H (2008) Insights into ligand selectivity in estrogen receptor isoforms: molecular dynamics simulations and binding free energy calculations. J Phys Chem B 112:2719–2726
Rastelli G, Rio AD, Degliesposti G, Sgobba M (2010) Fast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA. J Comput Chem 31:797–810
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23:3–25
Wu C, Wang Z, Lei H, Zhang W, Duan Y (2007) Dual binding modes of Congo red to amyloid protofibril surface observed in molecular dynamics simulations. J Am Chem Soc 129:1225–1232
Cooper JH (1974) Selective staining of amyloid as a function of amyloid composition and structure. Lab Invest 31:232–238
Klunk WE, Pettegrew JW, Abraham DJ (1989) Quantitative evaluation of congo red binding to amyloid-like proteins with a β-pleated sheet conformation. J Histochem Cytochem 37:1273–1281
Glenner GG (1980) Amyloid deposits and amyloidosis. N Engl J Med 302:1283–1292
Jin LW, Claborn KA, Kurimoto M, Geday MA, Maezawa I, Sohraby F, Estrada M, Kaminksy W, Kahr B (2003) Imaging linear birefringence and dichroism in cerebral amyloid pathologies. Proc Natl Acad Sci USA 100:15294–15298
Romhányi G (1971) Selective differentiation between amyloid and connective tissue structures based on the collagen specific topo-optical staining reaction with Congo red. Virchows Arch A Pathol Anat 354:209–222
Biancalana M, Makabe K, Koide A, Koide S (2009) Molecular mechanism of thioflavin-T binding to the surface of β-rich peptide self-assemblies. J Mol Biol 385:1052–1063
Childers WS, Mehta AK, Lu K, Lynn DG (2009) Templating molecular arrays in amyloid’s cross-β grooves. J Am Chem Soc 131:10165–10172
Maezawa I, Hong HS, Liu R, Wu CY, Cheng RH, Kung MP, Kung HF, Lam KS, Oddo S, LaFerla FM (2008) Congo red and thioflavin-T analogs detect Aβ oligomers. J Neurochem 104:457–468
Chimon S, Ishii Y (2005) Capturing intermediate structures of Alzheimer’s β-amyloid, Aβ (1–40), by solid-state NMR spectroscopy. J Am Chem Soc 127:13472–13473
Levine H 3rd (2005) Multiple ligand binding sites on Aβ (1–40) fibrils. Amyloid 12:5–14
Miura T, Yamamiya C, Sasaki M, Suzuki K, Takeuchi H (2002) Binding mode of Congo red to Alzheimer’s amyloid-β peptide studied by UV Raman spectroscopy. J Raman Spectrosc 33:530–535
Elhaddaoui A, Delacourte A, Turrell S (1993) Spectroscopic study of Congo red and thioflavin binding to amyloid-like proteins. J Mol Struct 294:115–118
Sajid J, Elhaddaoui A, Turrell S (1997) Investigation of the binding of Congo red to amyloid in Alzheimer’s diseased tissue. J Mol Struct 408:181–184
Pigorsch E, Elhaddaoui A, Turrell S (1994) Spectroscopic study of pH and solvent effects on the structure of Congo red and its binding mechanism to amyloid-like proteins. Spectrochim Acta A Mol Spectrosc 50:2145–2152
Esler WP, Stimson ER, Jennings JM, Vinters HV, Ghilardi JR, Lee JP, Mantyh PW, Maggio JE (2000) Alzheimer’s disease amyloid propagation by a template-dependent dock-lock mechanism. Biochemistry 39:6288–6295
Cannon MJ, Williams AD, Wetzel R, Myszka DG (2004) Kinetic analysis of β-amyloid fibril elongation. Anal Biochem 328:67–75
Ban T, Hoshino M, Takahashi S, Hamada D, Hasegawa K, Naiki H, Goto Y (2004) Direct observation of Aβ amyloid fibril growth and inhibition. J Mol Biol 344:757–767
Porat Y, Abramowitz A, Gazit E (2006) Inhibition of amyloid fibril formation by polyphenols: structural similarity and aromatic interactions as a common inhibition mechanism. Chem Biol Drug Des 67:27–37
Frid P, Anisimov SV, Popovic N (2007) Congo red and protein aggregation in neurodegenerative diseases. Brain Res Rev 53:135–160
Acknowledgments
The authors gratefully acknowledge the financial supports from Atomic Energy Council of Taiwan (Project number: ARA010203), National Science Council of Taiwan (Project numbers: 99-2221-E-027-022-MY3, 99-2221-E-027-037-MY2, and 99-2622-E-027-003-CC3), the Institute of Nuclear Energy Research of Taiwan (Project number: 1002001INER081), and National Taipei University of Technology and Taipei Medical University (Project number: NTUT-TMU-100-09).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 1115 kb)
Rights and permissions
About this article
Cite this article
Zhao, JH., Liu, HL., Elumalai, P. et al. Molecular modeling to investigate the binding of Congo red toward GNNQQNY protofibril and in silico virtual screening for the identification of new aggregation inhibitors. J Mol Model 19, 151–162 (2013). https://doi.org/10.1007/s00894-012-1532-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1532-y