Abstract
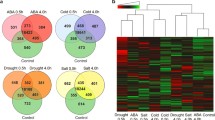
Genome-wide characterization of the Pohlia nutans transcriptome is essential for clarifying the role of stress-relevant genes in Antarctic moss adapting to the extreme polar environment. High-throughput Illumina sequencing was used to analyze the gene expression profile of P. nutans after cold treatment. A total of 93,488 unigenes, with an average length of 405 bp, were obtained. Gene annotation showed that 16,781 unigenes had significant similarity to known functional protein-coding genes, most of which were annotated using the GO, KOG and KEGG pathway databases. Global profiling of the differentially expressed genes revealed that 3,796 unigenes were significantly upregulated after cold treatment, while 1,405 unigenes were significantly downregulated. In addition, 816 receptor-like kinases and 1,309 transcription factors were identified from P. nutans. This overall survey of transcripts and stress-relevant genes can contribute to understanding the stress-resistance mechanism of Antarctic moss and will accelerate the practical exploitation of the genetic resources for this organism.




Similar content being viewed by others
References
Afoufa-Bastien D, Medici A, Jeauffre J, Coutos-Thévenot P, Lemoine R, Atanassova R, Laloi M (2010) The Vitis vinifera sugar transporter gene family: phylogenetic overview and macroarray expression profiling. BMC Plant Biol 10:245
Bhyan SB, Minami A, Kaneko Y, Suzuki S, Arakawa K, Sakata Y, Takezawa D (2012) Cold acclimation in the moss Physcomitrella patens involves abscisic acid-dependent signaling. J Plant Physiol 169(2):137–145
Blow N (2009) Transcriptomics: the digital generation. Nature 458:239–242
Bokhorst S, Huiskes A, Convey P, Aerts R (2007) The effect of environmental change on vascular plant and cryptogam communities from the Falkland Islands and the Maritime Antarctic. BMC Ecol 7:15
Chen ZZ, Xue CH, Zhu S, Zhou FF, Ling XB, Liu GP, Chen LB (2005) GoPipe: streamlined gene ontology annotation for batch anonymous sequences with statistics. Prog Biochem Biophys 32:187–191
Convey P, Stevens MI (2007) Antarctic Biodiversity. Science 317:1877–1878
Gish LA, Clark SE (2011) The RLK/Pelle family of kinases. Plant J 66:117–127
Hsiao YY, Chen YW, Huang SC, Pan ZJ, Fu CH, Chen WH, Tsai WC, Chen HH (2011) Gene discovery using next-generation pyrosequencing to develop ESTs for Phalaenopsis orchids. BMC Genomics 12:360
Huang W, Marth G (2008) EagleView: a genome assembly viewer for nextgeneration sequencing technologies. Genome Res 18:1538–1543
Hundertmark M, Hincha DK (2008) LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9:118
Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M (2010) KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 38:355–360
Lehti-Shiu MD, Zou C, Hanada K, Shiu SH (2009) Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol 150:12–26
Li XG, Wu HX, Southerton SG (2011) Transcriptome profiling of Pinus radiata juvenile wood with contrasting stiffness identifies putative candidate genes involved in microfibril orientation and cell wall mechanics. BMC Genomics 12:480
Libault M, Joshi T, Benedito VA, Xu D, Udvardi MK, Stacey G (2009) Legume transcription factor genes: what makes legumes so special? Plant Physiol 151:991–1001
Liu SH, Lee HS, Kang PS, Huang XH, Yim JH, Lee HK, Kim Il-C (2010) Complementary DNA library construction and expressed sequence tag analysis of an Arctic moss, Aulacomnium turgidum. Polar Biol 33:617–626
Lovelock CE, Jackson AE, Melick DR, Seppelt RD (1995) Reversible photoinhibition in Antarctic moss during freezing and thawing. Plant Physiol 109:955–961
Minami A, Nagao M, Ikegami K, Koshiba T, Arakawa K, Fujikawa S, Takezawa D (2005) Cold acclimation in bryophytes: low-temperature-induced freezing tolerance in Physcomitrella patens is associated with increases in expression levels of stress-related genes but not with increase in level of endogenous abscisic acid. Planta 220(3):414–423
Mizutani M, Sato F (2011) Unusual P450 reactions in plant secondary metabolism. Arch Biochem Biophys 507(1):194–203
Morgan-Kiss RM, Priscu JC, Pocock T, Gudynaite-Savitch L, Huner NP (2006) Adaptation and acclimation of photosynthetic microorganisms to permanently cold environments. Microbiol Mol Biol Rev 70:222–252
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Pearce DA (2008) Climate change and the microbiology of the Antarctic Peninsula region. Sci Prog 91:203–217
Peck LS, Clark MS, Clarke A et al (2005) Genomics: applications to Antarctic ecosystems. Polar Biol 28:351–365
Qi XH, Xu XW, Lin XJ, Zhang WJ, Chen XH (2012) Identification of differentially expressed genes in cucumber (Cucumis sativus L.) root under waterlogging stress by digital gene expression profile. Genomics 99:160–168
Ramel F, Sulmon C, Gouesbet G, Couée I (2009) Natural variation reveals relationships between pre-stress carbohydrate nutritional status and subsequent responses to xenobiotic and oxidative stress in Arabidopsis thaliana. Ann Bot 104(7):1323–1337
Reina-Bueno M, Argandoña M, Salvador M, Rodríguez-Moya J, Iglesias-Guerra F et al (2012) Role of trehalose in salinity and temperature tolerance in the model halophilic bacterium Chromohalobacter salexigens. PLoS ONE 7(3):e33587
Rensing SA, Lang D, Zimmer AD et al (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319:64–69
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16:276–277
Rodriguez MC, Petersen M, Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61:621–649
Rosenkranz R, Borodina T, Lehrach H, Himmelbauer H (2008) Characterizing the mouse ES cell transcriptome with Illumina sequencing. Genomics 92:187–194
Schulz MH, Zerbino DR, Vingron M, Birney E (2012) Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28:1086–1092
Schuster SC (2008) Next-generation sequencing transforms today’s biology. Nat Methods 5:16–18
Skotnicki ML, Ninham JA, Selkirk PM (2000) Genetic diversity, mutagenesis and dispersal of Antarctic mosses—a review of progress with molecular studies. Antarct Sci 12:363–373
Skotnicki ML, Mackenzie AM, Clements MA, Selkirk PM (2005) DNA sequencing and genetic diversity of the 18S–26S nuclear ribosomal internal transcribed spacers (ITS) in nine Antarctic moss species. Antarctic Sci 17:377–384
Sun MM, Li LH, Xie H, Ma RC, He YK (2007) Differentially expressed genes under cold acclimation in Physcomitrella patens. J Biochem Mol Biol 40(6):986–1001
Tibbett M, Sanders F, Cairney J (2002) Low-temperature-induced changes in trehalose, mannitol and arabitol associated with enhanced tolerance to freezing in ectomycorrhizal basidiomycetes (Hebeloma spp.). Mycorrhiza 12(5):249–255
Wang X, Yang P, Zhang X, Xu Y, Kuang T, Shen S, He Y (2009) Proteomic analysis of the cold stress response in the moss, Physcomitrella patens. Proteomics 9(19):4529–4538
Wang L, Feng Z, Wang X, Wang X, Zhang X (2010a) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138
Wang L, Li PH, Brutnell TP (2010b) Exploring plant transcriptomes using ultra high-throughput sequencing. Brief Funct Genomics 9:118–128
Wei WL, Qi XQ, Wang LH, Zhang YX, Hua W, Li DH, Lv HX, Zhang XR (2011) Characterization of the sesame (Sesamum indicum L.) global transcriptome using Illumina paired-end sequencing and development of EST-SSR markers. BMC Genomics 12:451
Wi SJ, Kim WT, Park KY (2006) Overexpression of carnation S-adenosylmethionine decarboxylase gene generates a broad-spectrum tolerance to abiotic stresses in transgenic tobacco plants. Plant Cell Rep 25:1111–1121
Xiang LX, He D, Dong WR, Zhang YW, Shao JZ (2010) Deep sequencing-based transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus reveals insight into the immune relevant genes in marine fish. BMC Genomics 11:472
Xiao L, Wang H, Wan P, Kuang T, He Y (2011) Genome-wide transcriptome analysis of gametophyte development in Physcomitrella patens. BMC Plant Biol 11:177
Yang T, Chaudhuri S, Yang L, Du L, Poovaiah BW (2010) A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J Biol Chem 285(10):7119–7126
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829
Zhang H, Jin JP, Tang L, Zhao Y, Gu XC, Gao G, Luo JC (2011) PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res 39:D1114–D1117
Zhou MQ, Shen C, Wu LH, Tang KX, Lin J (2011) CBF-dependent signaling pathway: a key responder to low temperature stress in plants. Crit Rev Biotechnol 31(2):186–192
Acknowledgments
We thank the Chinese National Human Genome Center (Shanghai) for their collaboration and support in the de novo assembly and annotation of the P. nutans transcriptome. We are very grateful for the critical comments and suggestions from the anonymous reviewers. This work was financially supported by the China National Natural Science Foundation (40906103 and 41206176) and Basic Scientific Research Foundation of The First Institute of Oceanography (SOA).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Atomi.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, S., Wang, N., Zhang, P. et al. Next-generation sequencing-based transcriptome profiling analysis of Pohlia nutans reveals insight into the stress-relevant genes in Antarctic moss. Extremophiles 17, 391–403 (2013). https://doi.org/10.1007/s00792-013-0528-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-013-0528-6