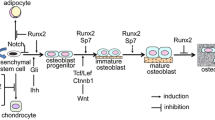
Abstract
Activating transcription factor 4 (ATF4) protein has a dual role in osteoblasts. It functions as a responder to stress to the endoplasmic reticulum (ER) as well as a transcription factor for bone formation. Little is known about molecular pathways that can potentially lead to stress-induced apoptosis or homeostasis of extracellular matrix (ECM) molecules. Based on microarray-derived mRNA expression data for mouse osteoblasts (MC3T3 E1 cells, clone 4), we analyzed the ER-stress responses in the presence of 10 nM Thapsigargin using two computational approaches: “Gene Set Enrichment Analysis (GSEA)” and “Ingenuity Pathways Analysis (IPA).” GSEA presented a strong linkage to an expression pattern observed in the responses to hypoxia, and IPA identified two molecular pathways: ATF4-unlinked connective tissue development and ATF4-linked organ morphology. Real-time polymerase chain reaction (PCR) and Western blot analyses validated eIF2α-driven translational regulation as well as ATF4-linked transcriptional activation of transcription factors and growth factors including FOS, FGF-9, and BMP-2. Consistent with the role of p38 MAPK in hypoxia, phosphorylation of p38 MAPK was activated in nonapoptotic osteoblasts under surviving ER stress. Furthermore, the level of phosphorylated PERK was elevated. These results support cross-talk between p38 MAPK and ER kinase, presenting a similarity to the responses to hypoxia as well as a pathway toward connective tissue development and organ morphology.
Similar content being viewed by others
References
Szegezdi E, Logue SE, Gorman AM, Samali A (2006) Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep 7:880–885
Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D (2006) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev 18:3066–3077
Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, Hanauer A, Karsenty G (2004) ATF4 is a substrate of RSK2 and essential regulator of osteoblast biology: implication for Coffi n-Lowry syndrome. Cell 117:387–398
Elefteriou F, Benson MD, Sowa H, Starbuck M, Liu X, Ron D, Parada LF, Karsenty G (2006) ATF4 mediation of NF1 functions in osteoblast reveals a nutritional basis for congenital skeletal dysplasiae. Cell Metab 4:441–451
Hamamura K, Yokota H (2007) Stress to endoplasmic reticulum of mouse osteoblasts induces apoptosis and transcriptional activation for bone remodeling. FEBS Lett 581:1769–1774
Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747–754
Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755–764
Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GWH, Beddington RSP, Mundlos S, Olsen BR, Selby PB, Owen MJ (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89:765–771
Wek RC, Jiang HY, Anthony TG (2006) Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 34:7–11
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Eber BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledgebased approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:12545–12550
Mayburd AL, Martlinez A, Sackett D, Liu H, Shih J, Tauler J, Avis I, Mulshine JL (2006) Ingenuity network-assisted transcription profiling: identification of a new pharmacologic mechanism for MK886. Clin Cancer Res 12:1820–1827
Vattem KM, Wek RC (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A 101:11269–11274
Dannenberg L, Edenberg HJ (2006) Epigenetics of gene expression in human hepatoma cells: expression profiling the response to inhibition of DNA methylation and histone deacetylation. BMC Genomics 7:181 (doi:10.1186/1471-2164-7-181)
Bouma GJ, Hart GT, Washburn LL, Recknagel AK, Eicher EM (2004) Using real time RT-PCR analysis to determine multiple gene expression patterns during XX and YY mouse fetal gonad development. Gene Expr Patterns 5:141–149
Mori K, Kitazawa R, Kondo T, Maeda S, Yamaguchi A, Kitazawa S (2006) Modulation of mouse RANKL gene expression by Runx2 and PKA pathway. J Cell Biochem 98:1629–1644
Wang D, Christensen K, Chawla K, Xiao G, Krebsbach PH, Franceschi RT (1999) Isolation and characterization of MC3T3-E1 preosteoblast subclones with distinct in vitro and in vivo differentiation/mineralization potential. J Bone Miner Res 14:893–903
Blais JD, Filipenko Bi VM, Harding HP, Ron D, Koumenis C, Wouters BJ, Bell JC (2004) Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol 24:7469–8211
Corazzari M, Lovat PE, Armstrong JL, Fimia GM, Hill DS, Birch-Machin M, Redfern CP, Piacentini M (2007) Targeting homeostatic mechanisms of endoplasmic reticulum stress to increase susceptibility of cancer cells to fenretinide-induced apoptosis: the role of stress proteins ERdj5 and ERp57. Br J Cancer 96:1062–1071
Herr I, Debatin KM (2001) Cellular stress response and apoptosis in cancer therapy. Blood 98:2603–2614
Seko Y, Takahashi N, Tobe K, Kadowaki T, Yazaki Y (1997) Hypoxia and hypoxia/reoxygenation activate p65PAK, p38 mitogen-activated protein kinase (MAPK), and stress-activated protein kinase (SAPK) in cultured rat cardiac myocytes. Biochem Biophys Res Commun 239:840–844
Steinmüller L, Cibelli G, Moll JR, Vinson C, Thiel G (2001) Regulation and composition of activator protein 1 (AP-1) transcription factors controlling collagenase and c-Jun promoter activities. Biochem J 360:599–607
Koumenis C, Naczki C, Koritzinsky M, Rastani S, Diehl A, Sonenberg N, Koromilas A, Wouters BG (2002) Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2α. Mol Cell Biol 22:7405–7416
Hu Y, Chan E, Wang SX, Li B (2003) Activation of p38 mitogenactivated protein kinase is required for osteoblast differentiation. Endocrinology 144:2068–2074
Bi M, Nacki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, Scheuner D, Kaufman RJ, Bell J, Ron D, Wouters BG, Koumenis C (2005) ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J 24:3470–3481
Ornitz DM, Itoh N (2001) Fibroblast growth factors. Genome Biol 2:3005.1–3005.12
Fakhry A, Ratisoontorn C, Vedhachalam C, Salhab I, Koyama E, Leboy P, Pacifici M, Kirschner RE, Nah HD (2005) Effects of FGF-2/-9 in calvarial bone cell cultures: differentiation stage-dependent mitogenic effect, inverse regulation of BMP-2 and noggin, and enhancement of osteogenic potential. Bone (NY) 36:254–266
Wozney JM (1992) The bone morphogenetic protein family and osteogenesis. Mol Reprod Dev 32:160–167
Nakase T, Yoshikawa H (2006) Potential roles of bone morphogenetic proteins (BMPs) in skeletal and regeneration. J Bone Miner Metab 24:425–433
Wagner EF (2002) Functions of AP1 (Fos/Jun) in bone development. Ann Rheum Dis 61:40–42
Inoue D, Kido S, Matsumoto T (2004) Transcriptional induction of FosB/ΔFosB gene by mechanical stress in osteoblasts. J Biol Chem 279:49795–49803
Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, Crombrugghe B (2002) The novel zinc fi nger-containing transcription factor Osterix is required for osteoblast differentiation and bone formation. Cell 108:17–29
Burge R, Dawson-Hughes B, Solomon DH, Wong JB, King A, Tosteson A (2007) Incidence and economic burden of osteoporosis-related fractures in the United States, 2005–2025. J Bone Miner Res 22:465–475
Author information
Authors and Affiliations
Corresponding author
Additional information
Electronic supplementary material The online version of this article (doi:10.1007/s00774-007-0825-1) contains supplementary material, which is available to authorized users.
Electronic supplementary material
About this article
Cite this article
Hamamura, K., Liu, Y. & Yokota, H. Microarray analysis of thapsigargin — induced stress to the endoplasmic reticulum of mouse osteoblasts. J Bone Miner Metab 26, 231–240 (2008). https://doi.org/10.1007/s00774-007-0825-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00774-007-0825-1