Abstract
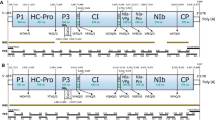
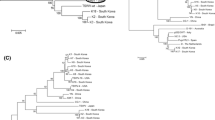
Plantago asiatica mosaic virus (PlAMV) is a member of the genus Potexvirus and has an exceptionally wide host range. It causes severe damage to lilies. Here we report on the complete nucleotide sequences of two new Japanese PlAMV isolates, one from the eudicot weed Viola grypoceras (PlAMV-Vi), and the other from the eudicot shrub Nandina domestica Thunb. (PlAMV-NJ). Their genomes contain five open reading frames (ORFs), which is characteristic of potexviruses. Surprisingly, the isolates showed only 76.0–78.0 % sequence identity with each other and with other PlAMV isolates, including isolates from Japanese lily and American nandina. Amino acid alignments of the replicase coding region encoded by ORF1 showed that the regions between the methyltransferase and helicase domains were less conserved than other regions, with several insertions and/or deletions. Phylogenetic analyses of the full-length nucleotide sequences revealed a moderate correlation between phylogenetic clustering and the original host plants of the PlAMV isolates. This study revealed the presence of two highly divergent PlAMV isolates in Japan.

Similar content being viewed by others
References
De La Torre CM, Qu F, Redinbaugh MG, Lewandowski DJ (2012) Biological and molecular characterization of a U.S. isolate of Hosta virus X. Phytopathology 102:1176–1181
Hammond J, Bampi D, Reinsel MD (2015) First report of Plantago asiatica mosaic virus in imported Asiatic and Oriental lilies (Lilium hybrids) in the United States. Plant Dis 99:292
Hughes PL, Harper F, Zimmerman MT, Scott SW (2005) Nandina mosaic virus is an isolate of Plantago asiatica mosaic virus. Eur J Plant Pathol 113:309–313
Komatsu K, Yamaji Y, Ozeki J, Hashimoto M, Kagiwada S, Takahashi S, Namba S (2008) Nucleotide sequence analysis of seven Japanese isolates of Plantago asiatica mosaic virus (PlAMV): a unique potexvirus with significantly high genomic and biological variability within the species. Arch Virol 153:193–198
Martelli GP, Adams MJ, Kreuze JF, Dolja VV (2007) Family Flexiviridae: a case study in virion and genome plasticity. Annu Rev Phytopathol 45:73–100
Pájtli É, Eke S, Palkovics L (2015) First report of the Plantago asiatica mosaic virus (PlAMV) incidence on Lilium sp. in Hungary. Plant Dis 99:1288
Parrella G, Greco B, Pasqualini A, Nappo AG (2015) Plantago asiatica mosaic virus found in protected crops of lily hybrids in Southern Italy. Plant Dis 99:1289
Solovyev AG, Novikov VK, Merits A, Savenkov EI, Zelentina DA, Tyulkina LG, Morozov SYu (1994) Genome characterization and taxonomy of Plantago asiatica mosaic potexvirus. J Gen Virol 75:259–267
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Yamaji Y, Kagiwada S, Nakabayashi H, Ugaki M, Namba S (2001) Complete nucleotide sequence of Tulip virus X (TVX-J): the border between species and strains within the genus Potexvirus. Arch Virol 146:2309–2320
Yusa A, Iwabuchi N, Koinuma H, Keima T, Neriya Y, Hashimoto M, Maejima K, Yamaji Y, Namba S (2016) Complete genome sequences of two hydrangea ringspot virus isolates from Japan. Genome Announc 4(2):e00022-16
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by a Grant-in-Aid for Young Scientists (B) (26850231) from the Japan Society for the Promotion of Science.
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with animals and human participants performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2016_3110_MOESM2_ESM.pptx
Fig. S1 Alignment of amino acid sequences of the replicase encoded by ORF1 in the PlAMV isolates, shown in Fig. 1, and closely related potexvirus species, TVX, and HdRSV. An alignment of the region between the methyltransferase and helicase domains of the replicase is shown. The numbers at the beginning of the alignment show the amino acid positions of the replicase (PPTX 360 kb)
Rights and permissions
About this article
Cite this article
Komatsu, K., Yamashita, K., Sugawara, K. et al. Complete genome sequences of two highly divergent Japanese isolates of Plantago asiatica mosaic virus. Arch Virol 162, 581–584 (2017). https://doi.org/10.1007/s00705-016-3110-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-3110-6