Abstract
Wood formation is an economically and environmentally important process and has played a significant role in the evolution of terrestrial plants. Despite its significance, the molecular underpinnings of the process are still poorly understood. We have previously shown that four lateral organ boundaries (LBD) transcription factors have important roles in the regulation of secondary (woody) growth with two (LBD1 and LBD4) involved in secondary phloem and ray cell development and two (LBD15 and LBD18) in secondary xylem formation. We studied gene copy number and variation in DNA and amino acid sequences of the four LBDs in a wide range of woody and herbaceous plant taxa with fully sequenced and annotated genomes. LBD1 showed the highest gene copy number across species, and gene copy number was strongly and significantly correlated with tangential ray width. The climbing vines, cucumber and grape, with wide multiseriate rays (>10 cells wide) showed the highest gene copy number. Because the growth habit of woody lianas like grape requires significant twisting and bending, it was suggested that the unlignified ray parenchyma cells likely facilitate stem flexibility and maintenance of xylem conductivity. We further demonstrate conservation of amino acids in LBD18 protein sequences specific to woody taxa downstream of the LBD domain. Neutrality tests showed evidence for strong purifying selection on these regions across various orders, indicating important functional roles in woody taxa. Additionally, structural modeling demonstrates that these regions have a significant impact on tertiary protein structure and thus are likely of significant functional importance.





Similar content being viewed by others
References
Alfieri FJ, Evert RF (1968) Seasonal development of the secondary phloem in Pinus. Amer J Bot 55:518–528
Angyalossy V, Pace MR, Lima AC (2015) Liana anatomy: a broad perspective on structural evolution of the vascular system. In: Schnitzer S, Bongers F, Burnham RJ, Putz FE (eds) Ecology of lianas. Wiley, Chichester, pp 251–287
Basson PW, Bierhorst DW (1967) An analysis of differential lateral growth in the stem of Bauhinia surinamensis. Bull Torrey Bot Club 94:404–411
Birol I, Raymond A, Jackman SD, Pleasance S, Coope R, Taylor GA, Saint Yuen MM, Keeling CI, Brand D, Vandervalk BP, Kirk H, Pandoh P, Moore RA, Zhao YJ, Mungall AJ, Jaquish B, Yanchuk A, Ritland C, Boyle B, Bousquet J, Ritland K, MacKay J, Bohlmann J, Jones SJM (2013) Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics 29:1492–1497
Bowers PM, Pellegrini M, Thompson MJ, Fierro J, Yeates TO, Eisenberg D (2004) Prolinks: a database of protein functional linkages derived from coevolution. Genome Biol 5:R35
Carlquist S (1985) Observations on functional wood histology of vines and lianas: vessel dimorphism, tracheids, vasicentric tracheids, narrow vessels, and parenchyma. Aliso 11:139–157
Carlquist S (1991) Anatomy of vine and liana stems: a review and synthesis. In: Putz FE, Mooney HA (eds) The biology of vines. Cambridge University Press, Cambridge, pp 53–71
Carlquist S (2001) Comparative wood anatomy: systematic, ecological, and evolutionary aspects of dicotyledon wood. Springer-Verlag, New York
Cavender-Bares J (2005) Impacts of freezing on long distance transport in woody plants. In: Holbrook NM, Zwieniecki M (eds) Vascular transport in plants. Elsevier Inc., Oxford, pp 401–424
Chase MW, Reveal JL (2009) A phylogenetic classification of the land plants to accompany APG III. Bot J Linn Soc 161:122–127
Craik CS, Rutter WJ, Fletterick R (1983) Splice junctions: association with variation in protein structure. Science 220:1125–1129
de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N (2006) ScanProsite: detection of prosite signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34:W362–W365
Doron-Faigenboim A, Stern A, Mayrose I, Bacharach E, Pupko T (2005) Selecton: a server for detecting evolutionary forces at a single amino-acid site. Bioinformatics 21:2101–2103
Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113
Fahn A (1990) Plant anatomy, 4th edn. Pergamon Press, Oxford
Fisher JB, Ewers FW (1989) Wound healing in stems of lianas after twisting and girdling injuries. Bot Gaz 150:251–265
Fitch R (1992) WinSTAT, the statistics program for Windows. Kalmia, Cambridge
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40:D1178–D1186
Gotmare V, Singh P, Tule BN (2000) Wild and cultivated species of cotton. CICR Technical Bulletin 5. Central Institute for Cotton Research, Nagpur
Groover AT (2005) What genes make a tree a tree? Trends Pl Sci 10:210–214
Groover AT, Nieminen K, Helariutta Y, Mansfield SD (2010). Wood formation in Populus. In: Genetics and Genomics of Populus, Springer, New York, pp 201–224
Hall BG (2011) Phylogenetic trees made easy: a how-to manual, 4th edn. Sinauer Associates, Sunderland
Hammond N (1991) Cuello: an early Maya community in Belize. Cambridge University Press, Cambridge
Hu Y, Zhang J, Jia H, Sosso D, Li T, Frommer WB, Yang B, White FF, Wang N, Jones JB (2014) Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc Natl Acad Sci USA 111:E521–E529
IPNI (2014) The International Plant Names Index. Available at: http://www.ipni.org/index.html, Accessed 26 Feb 2014
Iwakawa H, Ueno Y, Semiarti E, Onouchi H, Kojima S, Tsukaya H, Hasebe M, Soma T, Ikezaki M, Machida C, Machida Y (2002) The Asymmetric Leaves2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Pl Cell Physiol 43:467–478
Kempe A, Lautenschläger T, Lange A, Neinhuis C (2014) How to become a tree without wood—biomechanical analysis of the stem of Carica papaya L. Pl Biol (Stuttgart) 16:264–271
Kumar R, Choudhary V, Mishra S, Varma I, Mattiason B (2002) Adhesives and plastics based on soy protein products. Industr Crops Prod 16:155–172
Larson PR (1994) The vascular cambium: development and structure. Springer-Verlag, Berlin
Li Z, Zou L, Ye G, Xiong L, Ji Z, Zakria M, Hong N, Wang G, Chen G (2014) A potential disease susceptibility gene CsLOB of citrus is targeted by a major virulence effector PthA of Xanthomonas citri subsp. citri. Molec Pl 7:912–915
Lough TJ, Lucas WJ (2006) Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annual Rev Pl Biol 57:203–232
Majer C, Hochholdinger F (2011) Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci 16:47–52
Masrahi YS (2014) Ecological significance of wood anatomy in two lianas from arid southwestern Saudi Arabia. Saudi J Biol Sci 21:334–341
Matsumura Y, Iwakawa H, Machida Y, Machida C (2009) Characterization of genes in the I (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Pl J 58:525–537
Mellerowicz EJ, Baucher M, Sundberg B, Boerjan W (2001) Unravelling cell wall formation in the woody dicot stem. Pl Molec Biol 47:239–274
Neale D, Wegrzyn J, Stevens K, Zimin A, Puiu D, Crepeau M, Cardeno C, Koriabine M, Holtz-Morris A, Liechty J, Martinez-Garcia P, Vasquez-Gross H, Lin B, Zieve J, Dougherty W, Fuentes-Soriano S, Wu L-S, Gilbert D, Marcais G, Roberts M, Holt C, Yandell M, Davis J, Smith K, Dean J, Lorenz W, Whetten R, Sederoff R, Wheeler N, McGuire P, Main D, Loopstra C, Mockaitis K, deJong P, Yorke J, Salzberg S, Langley C (2014) Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol 15:R59
Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, Vicedomini R, Sahlin K, Sherwood E, Elfstrand M, Gramzow L, Holmberg K, Hallman J, Keech O, Klasson L, Koriabine M, Kucukoglu M, Kaller M, Luthman J, Lysholm F, Niittyla T, Olson A, Rilakovic N, Ritland C, Rossello JA, Sena J, Svensson T, Talavera-Lopez C, Theissen G, Tuominen H, Vanneste K, Wu ZQ, Zhang B, Zerbe P, Arvestad L, Bhalerao R, Bohlmann J, Bousquet J, Gil RG, Hvidsten TR, de Jong P, MacKay J, Morgante M, Ritland K, Sundberg B, Thompson SL, Van de Peer Y, Andersson B, Nilsson O, Ingvarsson PK, Lundeberg J, Jansson S (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497:579–584
Parker J, Tsagkogeorga G, Cotton JA, Liu Y, Provero P, Stupka E, Rossiter SJ (2013) Genome-wide signatures of convergent evolution in echolocating mammals. Nature 502:228–231
Plomion C, Leprovost G, Stokes A (2001) Wood formation in trees. Pl Physiol 127:1513–1523
Poorter L, McDonald I, Alarcon A, Fichtler E, Licona JC, Pena-Claros M, Sterck F, Villegas Z, Sass-Klaassen U (2010) The importance of wood traits and hydraulic conductance for the performance and life history strategies of 42 rainforest tree species. New Phytol 185:481–492
Putz F, Holbrook N (1991) Biomechanical studies of vines. In: Putz FE, Mooney HA (eds) The biology of vines, Cambridge University Press, Cambridge, pp 73–97
Růžička K, Ursache R, Hejátko J, Helariutta Y (2015) Xylem development—from the cradle to the grave. New Phytol 207:519–535
Schweingruber F, Landolt W (2005) The xylem database. Available at: http://www.wsl.ch/dendropro/xylemdb/, Accessed 14 Oct 2014
Schweingruber FH, Börner A, Schulze E-D (2007) Atlas of woody plant stems: evolution, structure, and environmental modifications. Springer, Berlin-Heidelberg
Scofield S, Murray JAH (2006) KNOX gene function in plant stem cell niches. Pl Molec Biol 60:929–946
Sedjo RA (2001) The role of forest plantations in the world’s future timber supply. Forest Chron 77:221–225
Shuai B, Reynaga-Pena CG, Springer PS (2002) The lateral organ boundaries gene defines a novel, plant-specific gene family. Pl Physiol 129:747–761
Soyano T, Thitamadee S, Machida Y, Chua NH (2008) Asymmetric Leaves2-Like19/Lateral Organ Boundaries Domain30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Pl Cell 20:3359–3373
Spicer R, Groover A (2010) Evolution of development of vascular cambia and secondary growth. New Phytol 186:577–592
Spicer R (2014) Symplasmic networks in secondary vascular tissues: parenchyma distribution and activity supporting long-distance transport. J Exp Bot 65:1829–1848
Stern A, Doron-Faigenboim A, Erez E, Martz E, Bacharach E, Pupko T (2007) Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res 35:506–511
Stevens PF (2001) Angiosperm Phylogeny Website, version 13. Available at: http://www.mobot.org/MOBOT/research/APweb/, Accessed 14 October 2014
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molec Biol Evol 28:2731–2739
van der Schoot C, van Bel AJ (1989) Architecture of the internodal xylem of tomato (Solanum lycopersicum) with reference to longitudinal and lateral transfer. Am J Bot 76:487–503
Warren RL, Keeling CI, Yuen MMS, Raymond A, Taylor GA, Vandervalk BP, Mohamadi H, Paulino D, Chiu R, Jackman SD, Robertson G, Yang C, Boyle B, Hoffmann M, Weigel D, Nelson DR, Ritland C, Isabel N, Jaquish B, Yanchuk A, Bousquet J, Jones SJM, MacKay J, Birol I, Bohlmann J (2015) Improved white spruce (Picea glauca) genome assemblies and annotation of large gene families of conifer terpenoid and phenolic defense metabolism. Pl J 83:189–212
Wegrzyn JL, Liechty JD, Stevens KA, Wu LS, Loopstra CA, Vasquez-Gross H, Dougherty WM, Lin BY, Zieve JJ, Martinez-Garcia PJ, Holt C, Yandell M, Zimin A, Yorke JA, Crepeau M, Puiu D, Salzberg SL, de Jong P, Mockaitis K, Main D, Langley CH, Neale DB (2014) Unique features of the loblolly pine (Pinus taeda L.) megagenome revealed through sequence annotation. Genetics 196:891–909
Wheeler EA (2004-onwards) InsideWood Website. Available at: http://insidewood.lib.ncsu.edu/search, Accessed 5 May 2015
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8
Ye J, McGinnis S, Madden TL (2006) BLAST: improvements for better sequence analysis. Nucleic Acids Res 34:W6–W9
Yordanov YS, Busov V (2011) Boundary genes in regulation and evolution of secondary growth. Plant Signal Behav 6:688–690
Yordanov YS, Regan S, Busov V (2010) Members of the lateral organ boundaries domain transcription factor family are involved in the regulation of secondary growth in Populus. Pl Cell 22:3662–3677
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40
Zhang J, Elo A, Helariutta Y (2011) Arabidopsis as a model for wood formation. Curr Opinion Biotechnol 22:293–299
Zimin A, Stevens KA, Crepeau M, Holtz-Morris A, Koriabine M, Marcais G, Puiu D, Roberts M, Wegrzyn JL, de Jong PJ, Neale DB, Salzberg SL, Yorke JA, Langley CH (2014) Sequencing and assembly of the 22-Gb loblolly pine genome. Genetics 196:875–890
Acknowledgments
Funding for the project was provided by the USDA-NIFA grant (2012-67013-19389) to VB, YY and OG. We would also like to thank the Biochemistry and Molecular Biology program at Michigan Technological University (MTU) which supported the research for one semester. Additional support was made available by MTU’s Ecosystem Science and Biotech Research Center. We would like to thank for the very constructive comments from two anonymous reviewers.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Funding
This study was funded by the USDA-NIFA grant (grant number 2012-67013-19389).
Additional information
Handling editor: Günter Theißen.
Electronic supplementary material
Below is the link to the electronic supplementary material.
606_2015_1272_MOESM1_ESM.pdf
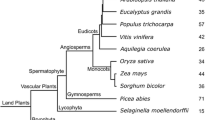
Neighbor-Joining tree based on complete protein sequences of all LBD genes from Arabidopsis thaliana and Brassica rapa. LBD homologs and gene copy numbers for each species are determined by the position in the phylogenetic tree relative to Arabidopsis thaliana LBD genes (PDF 35 kb)
606_2015_1272_MOESM2_ESM.pdf
Neighbor-Joining tree based on complete protein sequences of LBD1 genes. Bootstrap values above 50 and genes from woody species are highlighted in yellow (PDF 394 kb)
606_2015_1272_MOESM3_ESM.pdf
Neighbor-Joining tree based on complete protein sequences of LBD4 genes. Bootstrap values above 50 and genes from woody species are highlighted in yellow (PDF 277 kb)
606_2015_1272_MOESM4_ESM.pdf
Neighbor-Joining tree based on complete protein sequences of LBD15 genes. Bootstrap values above 50 and genes from woody species are highlighted in yellow (PDF 288 kb)
606_2015_1272_MOESM5_ESM.pdf
Neighbor-Joining tree based on complete protein sequences of LBD18 genes. Bootstrap values above 50 and genes from woody species are highlighted in yellow (PDF 47 kb)
606_2015_1272_MOESM6_ESM.pdf
Neighbor-Joining tree based on complete DNA coding sequences of LBD18 genes. Bootstrap values above 50 and genes from woody species are highlighted in yellow (PDF 47 kb)
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. Neighbor-Joining tree based on complete protein sequences of all LBD genes from Arabidopsis thaliana and Brassica rapa. LBD homologs and gene copy numbers for each species are determined by the position in the phylogenetic tree relative to Arabidopsis thaliana LBD genes.
Online Resource 2. Neighbor-Joining tree based on complete protein sequences of LBD1 genes. Bootstrap values above 50 and genes from woody species are highlighted in yellow.
Online Resource 3. Neighbor-Joining tree based on complete protein sequences of LBD4 genes. Bootstrap values above 50 and genes from woody species are highlighted in yellow.
Online Resource 4. Neighbor-Joining tree based on complete protein sequences of LBD15 genes. Bootstrap values above 50 and genes from woody species are highlighted in yellow.
Online Resource 5. Neighbor-Joining tree based on complete protein sequences of LBD18 genes. Bootstrap values above 50 and genes from woody species are highlighted in yellow.
Online Resource 6. Neighbor-Joining tree based on complete DNA coding sequences of LBD18 genes. Bootstrap values above 50 and genes from woody species are highlighted in yellow.
Rights and permissions
About this article
Cite this article
Bdeir, R., Busov, V., Yordanov, Y. et al. Gene dosage effects and signatures of purifying selection in lateral organ boundaries domain (LBD) genes LBD1 and LBD18. Plant Syst Evol 302, 433–445 (2016). https://doi.org/10.1007/s00606-015-1272-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-015-1272-4