Abstract
The new COVID-19 emerged in a town in China named Wuhan in December 2019, and since then, this deadly virus has infected 324 million people worldwide and caused 5.53 million deaths by January 2022. Because of the rapid spread of this pandemic, different countries are facing the problem of a shortage of resources, such as medical test kits and ventilators, as the number of cases increased uncontrollably. Therefore, developing a readily available, low-priced, and automated approach for COVID-19 identification is the need of the hour. The proposed study uses chest radiography images (CRIs) such as X-rays and computed tomography (CTs) to detect chest infections, as these modalities contain important information about chest infections. This research introduces a novel hybrid deep learning model named Lightweight ResGRU that uses residual blocks and a bidirectional gated recurrent unit to diagnose non-COVID and COVID-19 infections using pre-processed CRIs. Lightweight ResGRU is used for multi-modal two-class classification (normal and COVID-19), three-class classification (normal, COVID-19, and viral pneumonia), four-class classification (normal, COVID-19, viral pneumonia, and bacterial pneumonia), and COVID-19 severity types’ classification (i.e., atypical appearance, indeterminate appearance, typical appearance, and negative for pneumonia). The proposed architecture achieved f-measure of 99.0%, 98.4%, 91.0%, and 80.5% for two-class, three-class, four-class, and COVID-19 severity level classifications, respectively, on unseen data. A large dataset is created by combining and changing different publicly available datasets. The results prove that radiologists can adopt this method to screen chest infections where test kits are limited.
Similar content being viewed by others
1 Introduction
The "Severe Acute Respiratory Syndrome Coronavirus-2" (SARS-CoV-2), which causes COVID-19 infection, started in Wuhan, China, at the end of December 2019 and was transmitted worldwide in a few days. This infectious disease was initially named SARS-CoV-2, but the World Health Organization (WHO) renamed it COVID-19 [1, 2]. In January 2020, WHO declared COVID-19 a global health issue [1]. Later, on March 11, 2020, this disease was declared a pandemic by WHO [3]. The contagious disease infected 324 million people globally. It caused 5.53 million deaths till January 15, 2022 [4]. Common symptoms that are observed in different patients are fever, sour throat, throat swelling, sneezing, cough, body pain, headache, breathing issues, and chest infections [5]. Various countries are fighting to control this pandemic by early detection of this disease using manual laboratory tests. However, third-world countries like Pakistan are facing a scarcity of resources to diagnose this infection. Therefore, there is a need for low-cost, automatic COVID-19 detection mechanisms that can use medical imaging to identify and predict COVID-19 and its severity level.
The most common method for detecting COVID-19 infection is a nucleic acid analysis using a real-time “Reverse-Transcription Polymerase Chain Reaction” (RT-PCR). This method is slow, and it takes a minimum of one day to get results. The RT-PCR method has a low sensitivity of around 60%–70% [6]. The test has a high false-negative rate (FNR), which may cause the virus to spread quickly and the patient to receive incorrect treatment [7]. X-rays and CT are effective screening methods because they detect changes in the lungs before clinical symptoms of COVID-19 appear. These are also useful in detecting early lesions and damage in the lungs, and their results are better and more accurate than manual lab tests [6]. CRIs-based recognition systems have several advantages over regular blood and RT-PCR examinations (these methods are rapid, inexpensive, and require less human resources than other manual methods (i.e., RT-PCR)). According to the findings, CT scans of infected lungs are more sensitive in detecting COVID-19 pneumonia, even in asymptomatic patients. It has better performance in detecting infection when compared with RT-PCR [8, 9]. Thus, the physical testing methods should be substituted with an automated diagnostic system for COVID-19. There are X-rays and CTs of healthy people and COVID-19 infected people that are openly accessible, which facilitate researchers to inspect conceivable features for COVID-19 detection [10].
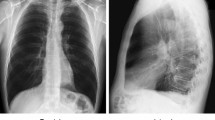
Medical imaging (e.g., X-rays and CTs) plays a vital role in diagnosing and treating COVID-19 chest infection. A chest X-ray is a standard radiography imaging type used to detect COVID-19 infection. It helps to identify the space around the lungs filled with fluid (blood or pus) or other abnormalities. In contrast, computed tomography (CT) is a specialized CRI that uses X-rays to form 3D images of the lungs. Though suspicious findings on these images, along with the patient's other medical histories, are useful clues for COVID-19 damage prediction. According to some studies, abnormalities have been reported in CRIs even before the scientific characterization of COVID-19 [11]. For example, COVID-19 patients have opacities in the right infrahilar area in the lungs, as presented by Kong et al. [12]. Therefore, CRIs analysis can provide proper guidance and direction to medical specialists and radiologists to understand the disease and examine clinical challenges in a low-cost and time-saving manner, especially in countries with a deficiency of inspection testing kits. Chest radiography images of the lungs are shown in Fig. 1.
The X-ray appearance of COVID-19 has been classified into four severity categories by the Radiological Society of North America (RSNA). They classified these severity levels into atypical, indeterminate, typical, and negative for pneumonia based on the grading of this infectious disease. Atypical appearance has lung abnormalities with a low-level infection, which can be consistent with other lung infections. Furthermore, a few symptoms of COVID-19 lung infection are present in an indeterminate appearance. The typical appearance has high abnormalities suspicious of COVID-19 infection. While negative for pneumonia type has no lung infection, it may be an asymptomatic COVID-19 infection [13, 14]. The chest X-rays of different severity levels of COVID-19 are depicted in Fig. 2.
Various generations of machine-learning methods, such as traditional deep neural networks and spiking neural networks (SNNs) [15], allow for breakthrough progress in various fields, including image processing. Deep neural learning (DL) has the potential to predict and identify distinct kinds of lung pneumonia because of its recent success in medical image analysis [16]. The core challenge is to predict COVID-19 pneumonia with low-cost and accurate detection methods with high sensitivity. A convolutional neural network (CNN) is a standard method of deep learning (DL) [17] that plays a significant role in medical imaging examinations, speech recognition, natural language processing (NLP), and audio recognition. It has an end-to-end learning process that automatically learns useful data features. Using these learned patterns, it builds a model that takes input and classifies data as accurately as possible.
This research paper presents a novel and hybrid architecture named Lightweight ResGRU. The proposed architecture uses residual blocks (RB) based on 2-dimensional CNN, pooling layers, dense layers, batch normalization layers, and a bidirectional gated recurrent unit (Bi-GRU) for the prediction of non-COVID chest infections (i.e., bacterial and viral) and COVID-19 infection and its different severity levels. The residual block is the building block in the proposed architecture because it speeds up model training and propagates lower-level features via skip connections [18]. Bi-GRU is a sequential processing model, a type of recurrent neural network (RNN). It is used in the architecture to compute the dependencies and connections between input data features [19], which improves classification accuracy. It also facilitates addressing the issue of intra-class similarities (i.e., similarities between images of different classes), as it keeps the information of previous and forward images of a current image in the data and predicts a class based on this information. The classification is categorized in terms of two-class multi-modal classification (normal and COVID-19), which uses X-rays and CTs for the training of the model, three-class classification (normal, COVID-19, and viral pneumonia), four-class classification (normal, COVID-19, viral pneumonia, and bacterial pneumonia), and SARS-CoV-2 severity levels’ classification (negative for pneumonia, atypical, indeterminate, and typical). Lightweight ResGRU uses different benchmark datasets [20]–[23] that contain X-rays and CTs of healthy and non-healthy (patients suffering from chest infections) for training, validation, testing, and external cohort validation (cross-dataset evaluation). Accuracy, precision, sensitivity, specificity, f-measure, false-negative rate (FNR), false-positive rate (FPR), and confusion matrix are used to evaluate the proposed system. The links to datasets and source codes are available at https://github.com/Mughees-Ahmad/Lightweight_ResGRU. The following are the main contributions of the paper:
-
A novel and hybrid model is showcased that combines the properties of residual blocks and bidirectional gated recurrent unit (Bi-GRU) for the early detection of chest infection with low false-negative rate and false-positive rate.
-
A lightweight approach is proposed in which skip connections are used to jump over some network layers, minimizing the number of parameters and making the model lightweight.
-
A larger dataset is curated and used for model training to improve generalizability and prevent the problem of model over-fitting. An external cohort/cross-dataset is also used to assess the performance of the suggested model on an external dataset obtained from a different source.
-
Multi-modal CRIs (X-Rays and CTs) are used to train the model for binary classification (normal and COVID-19). Thus, radiologists and medical experts can use either X-ray or CT to screen for COVID-19 pneumonia.
-
The suggested model predicts the distinct severity levels of COVID-19 (i.e., negative for pneumonia, atypical, indeterminate, and typical) proposed by RSNA, which, to the best of our knowledge, has not been published by any research study.
The rest of this article is divided into the following sections: After the introduction section, Sect. 2 will outline a literature survey of existing studies. Section 3 describes the datasets' details and the proposed system's complete architecture. Experimental setups are described in Sect. 4. The results of the proposed study are presented in Sect. 5. Section 6 displays the model's performance on the cross-dataset. Section 7 discusses and analyzes Lightweight ResGRU and its comparison with existing research studies. Finally, the conclusion of the research is presented in Sect. 8.
2 Literature survey
COVID-19 diagnosis using DL has become a well-established research tool since it became a global pandemic. Excellent deep learning-based research efforts that detect and identify COVID-19 in X-ray and CT scan images were discovered. Despite their impressive results, CNN techniques do not replace traditional testing methods. These techniques are helpful when combined with traditional testing procedures, but much more research is needed before they can be used practically [6]. A broad group of researchers and scientists are collaborating to develop accurate and reliable COVID-19 diagnosis methods based on deep learning.
Since 2020, many scholars have suggested different methods for COVID-19 detection using CRIs. Most scientists have recommended DL approaches for COVID-19 prediction. Several researchers focused on COVID and non-COVID binary classification [24,25,26,27,28]. However, many researchers have worked on three-class classification (normal, COVID-19, and pneumonia) as well [7, 29,30,31,32,33,34]. However, fewer focused on four-class classification (normal, COVID-19 pneumonia, bacterial pneumonia, and viral pneumonia) [6, 35].
Ardakani et al. [24] used ten advanced pre-trained deep learning models on a custom dataset of 1020 CT slices in their research to differentiate between COVID-19 and normal CRIs. Out of 10 CNN algorithms, Resnet-101 achieved an AUC of 0.994 (sensitivity, 100%; specificity, 99.02%; accuracy, 99.51%). Shibly et al. [25] introduced a framework comprised of VGG-16 and Faster R-CNN, which achieved a classification accuracy of 97.36%, 97.65% sensitivity, and 99.28% precision. This study made use of a dataset containing 19,250 X-ray images. Javor et al. [26] suggested a machine-learning algorithm with a simple architecture that attained an overall accuracy of 95.6% on an unseen test set of 90 X-ray images. The overall dataset contains 6868 X-ray images. Wang et al. [27] used CT images with DL techniques to monitor the COVID-19 infection in the lungs. They modified the Inception pre-trained model and used transfer learning to design an algorithm for COVID-19 classification. Their experiments achieved 89.5% accuracy. They externally validated their model and attained an accuracy of 79.3%. Ismael et al. [28] used different pre-trained CNN architectures for feature extraction and support vector machine (SVM) for binary classification (COVID and normal) by using a dataset of 380 X-ray images of the lungs. ResNet50 and SVM with a linear kernel achieved the highest accuracy score of 94.7% among different pre-trained models.
Das et al. [7] used VGG-16 and Resnet-50 in their study. VGG-16 got a high accuracy of 97.67% for three-class classification (COVID-19, pneumonia, and normal). A dataset of 2905 X-ray images was used for the training and validation of the model. In the research study [29], Dash et al. used a fine-tuned pre-trained model with chest radiography images. They proposed a unique framework by removing fully connected layers of VGG-16 and placing a new simplified, fully connected layer assigned with random weight. As a result, they achieved 97.1% accuracy, 99.2% sensitivity, and 99.6% specificity. Ozturk et al. [30] presented a deep learning framework named DarkCovidNet that contains seventeen convolutional layers (with a different number of filters in every layer), which used an overall of 1127 X-ray images. Their model attained an accuracy of 98.08% and 87.02% for binary classification and three-class classification (COVID-19, no-finding, and pneumonia), respectively. Islam et al. [31] proposed a hybrid architecture that contained CNN for feature extraction and long short-term memory (LSTM) for feature classification over a dataset consisting of 4575 X-ray images of the three-class problem. Their framework reached an accuracy of 99.4%. Demir et al. [32] presented a DL model (DeepCoroNet) created by convolutional layers as feature extractors and used the LSTM layer for classification. This model accomplished an overall accuracy of 100% on the three-class problem using 1,061 chest X-ray (CX) images. Turkoglu et al. [33] suggested a model named COVIDetectioNet, which consists of AlexNet for feature extraction, the Relief algorithm for the selection of useful features, and then classified those selected features using SVM. They achieved an overall accuracy of 99.18% for the three-class (COVID, normal, and pneumonia) classification. In the research study [34], Luz et al. used different versions of the EfficeintNet deep learning model for the three-class (COVID, normal, and pneumonia) problem by using a publically available dataset named COVIDx containing 13,569 chest X-rays. EfficeintNet B3-X outperformed all other versions with an accuracy of 93.9%.
Elkorany et al. [35] proposed a DL model (COVIDetection-Net) in which Squeezenet and Shufflenet models are used for feature extraction, and a multiclass support vector machine (MSVM) is used for classification. 1200 X-ray images are used in this study. The architecture attained an accuracy of 100%, 99.72%, and 94.44% for binary, three-class, and four-class classification, respectively. Hussain et al. [6] introduced a novel deep learning algorithm, CoroDet, which comprises 22 deep layers-based architecture. This study used X-ray and CT CRIs for model training and attained an accuracy of 99.1%, 94.2%, and 91.2% for two-class, three-class, and four-class classifications. 2,100 X-rays are used with training, validation, and testing split with a ratio of 80:10:10.
Because of its importance, various ML techniques have been presented to diagnose SARS-CoV-2 (COVID-19). Most researchers have focused on two-class (normal and COVID-19) and three-class (normal, COVID-19, and pneumonia) classifications. While less concentrated on the four-class (normal, COVID-19 pneumonia, bacterial pneumonia, and viral pneumonia) classification. Researchers have used a limited and small amount of data for network training, which can lead to the over-fitting of the model [36]. Furthermore, most researchers used a two-split (training and validation sets) approach for model training and testing, which may lead to biased high accuracy results [37]. Most of them did not use multi-modalities (X-rays and CTs) for model training and inference in their studies. Hence, the limitations of the existing advanced studies have encouraged us to develop an efficient technique for detecting different types of chest infections, including SARS-CoV-2. The study aims to differentiate between different severity levels of COVID-19 (negative for pneumonia, atypical, indeterminate, and typical), which, to the best of our knowledge, has not been published yet. The proposed method aims to provide a hybrid deep learning (DL) method based on multi-modal images that can be a cost-effective and novel prediction mechanism for this virus. This study can distinguish between COVID-19 chest infection and other lung infections (bacterial or viral), and it uses a large dataset for model training to enhance the generalizability of the system. Therefore, the proposed model can help doctors to determine the type of chest infection more accurately and quickly without requiring extensive physical examinations.
3 Methodology
This section defines the complete details of datasets and discusses the Lightweight ResGRU for detecting COVID-19 using CRIs, i.e., X-rays and CTs, gathered from different resources. The residual block is the building block of the proposed model because it uses skip connections and connects the activation of a layer to a deeper layer in a neural network, resulting in faster training of the model. Later, the proposed model also uses the bidirectional gated recurrent unit (Bi-GRU) to compute the dependency and connection of the features of the middle layer's output of the residual units. It associates the features of these intermediate layers to the last fully connected network for classification, which results in improved classification accuracy.
The workflow diagram of our presented model, Lightweight ResGRU, is shown in Fig. 3. The diagram provides a general outline of the proposed method from beginning to end. The detail of each step of the model Lightweight ResGRU is explained in the following sub-sections.
3.1 Dataset creation
In this research study, we started by generating a large dataset on COVID-19 prediction by using different standard datasets that are publicly available. The dataset is arranged by integrating and altering four different publicly available benchmark repositories on the internet. These different dataset repositories named Chest Radiography Database contains (normal: 10,192, COVID-19: 3,616, viral pneumonia: 1345, and bacterial pneumonia: 6012) X-ray images [21], “SARS-CoV-2 Ct-Scan” dataset consists (normal: 1229, and COVID-19: 1252) CT images [23], and a “Curated Dataset for COVID-19 Posterior-Anterior Chest Radiography Images (X-Rays)” which is used as an external cohort [38] to test the model performance on an external source dataset comprises (normal: 3270, COVID-19: 1281, viral pneumonia: 1656, and bacterial pneumonia: 3001) X-ray images [20], SIIM-FISABIO-RSNA COVID-19 Detection contains (atypical appearance: 483, indeterminate appearance: 1108, typical appearance: 3007, and negative for pneumonia: 1736) X-ray images [22].
After combining the datasets, the combined dataset is divided into four datasets i.e., a two-class classification dataset (normal and COVID-19) that contains multi-modal CRIs (X-rays and CTs), a three-class classification dataset (normal, COVID-19, and viral pneumonia), a four-class classification dataset, and a four-class severity level classification (negative for pneumonia, atypical, indeterminate, and typical).
3.1.1 Chest infection prediction dataset
The training dataset is organized by gathering and modifying two datasets, i.e., the Chest Radiography Database [21] and the SARS-CoV-2 Ct-Scan Dataset [23]. The details of the images in each class are described in Tables 1, 2, and 3. Multi-modal CRIs are utilized, and the model training for binary classification makes use of both X-rays and CT images, as shown in Table 3. The four sample images from the dataset are shown in Fig. 4.
3.1.2 COVID-19 severity detection dataset
The model also predicts the different severity levels of COVID-19. For this purpose, the “SIIM-FISABIO-RSNA COVID-19 Detection” dataset [22] is utilized for the training of the model. This dataset is made up of two independent source datasets, BIMCV-COVID-19 data [39] and MIDRC-RECORD data [40]. The dataset initially contained 7268 images of four different classes (atypical appearance, indeterminate appearance, typical appearance, and negative for pneumonia). Data augmentation is used to balance the dataset across different classes. The basic augmentation techniques (horizontal flip, zooming, and brightness) are used in the original dataset. The detail of the augmented dataset is explained in Table 4.
3.2 External cohort: for external/cross-dataset validation
The model's performance is evaluated against an external cohort to reduce the possibility of biased performance toward the training data [38]. The “Curated Dataset for COVID-19 Posterior-Anterior Chest Radiography Images (X-Rays)” was used as an external cohort [38] to check the generalizability of the model on an external dataset (normal: 3270, COVID-19: 1281, viral pneumonia: 1656, and bacterial pneumonia: 3001) [20]. A subset (i.e., 2700 random images) of this dataset is used as a cross-dataset to assess the performance of the proposed model. The detail of the dataset is described in Table 5.
3.3 Data pre-processing
This section describes the detail of the procedures used in the preprocessing. The preprocessing techniques are explained in the following sub-sections.
3.3.1 Resizing and normalization
First, the input images are resized to 224 * 224 resolution before being fed to the model to decrease the number of parameters (weights and biases) and, as a result, make the model simpler. Second, normalization is used in deep learning architectures to stabilize model training while accelerating training [41]. The image pixel values have been adjusted to the range [0–1] from [0–255] by dividing every pixel by 255. The images used in this study are grayscale.
3.3.2 Data augmentation
Deep learning architectures require a large dataset to effectively train the model and achieve better results [42]. Data augmentation is a technique to increase the amount of dataset artificially while preserving labels. The data augmentation applied to some classes which had fewer cases. A set of transformations were applied to the original dataset, including horizontal flip, zooming up by 20%, and brightening by 15%. The augmented samples of the dataset are displayed in Fig. 5.
3.4 Lightweight ResGRU: our proposed model
In this section, the proposed hybrid Lightweight ResGRU for the detection of COVID-19 and non-COVID-19 infections by using CRIs is described. The proposed model contains different components: residual block, batch normalization layer, pooling layer, flatten layer, dense layer, time-distributed layer, Bi-GRU, and activation functions, as shown in Fig. 6. The details of the different modules of the model are described in the following sub-sections.
3.4.1 Batch normalization layer
After the input layer, every two successive residual blocks and a dense layer, the batch normalization layer is applied. The proposed model has five batch normalization layers. The batch normalization layer standardizes the inputs of a layer for each batch before feeding them to the next layer in the network. It accelerates and stabilizes the DNN training process [43]. Batch normalization is usually applied after the activation function, which yields better results.
Let μb, and σ2b represent the batch's mean and variance, as given in Eqs. (1 and 2). Normalize the layer inputs using the batch statistics i.e., mean and variance, and then standardize the hidden units by scaling and shifting using the learned scaling parameters.
whereas, Eq. (3) shows the batch normalization formula, and Eq. (4) shows scaling and shifting using the learned scaling parameter γ and shift parameter β.
3.4.2 Residual learning block
In the proposed model, residual blocks are used after the input layer to tackle the network's vanishing gradient problem [18]. Residual learning facilitates faster training of the model and gives comparatively better results than other DNNs. The central idea is to add a skip connection that connects the activation of a layer deeper in the network, which means it flows the information of one layer and feeds about 2–3 layers deeper in the DNN. Thus, in the residual block, a direct link skips 2–3 layers in-between in a DNN, as shown in Fig. 7.
Residual Learning: A building block [17]
Let H(x) denotes the residual mapping where x is the input. Moreover, the stacked nonlinear layers use another function, F(x): = H(x) − x. Then the residual mapping is calculated as F(x) + x. Residual mapping H(x) can be easily optimized compared to the original mapping F(x). A neural network with skip connections is shown in Fig. 7.
Hence, residual blocks are based on convolution operation. The convolution operation with an image (X) and mask (M) is defined in Eq. (7).
Here, * denotes the convolution operation being used in the residual learning block. The window or mask (M) slides over the image (X) with a certain stride value. The model contains six residual blocks in total.
3.4.3 Pooling layer and dropout layer
The max pooling layer is applied frequently in the Lightweight ResGRU, which shrinks the image size and reduces the network's complexity. The max pooling layer stops over-fitting and accelerates the model's training. It proceeds with the maximum value of the feature map (F) under the mask (M). The hyper-parameters required for the max-pooling layer are a mask (M) and stride (S), as depicted in Eq. (8).
where ‘S’ is stride, and x and y are rows and columns of the input image, respectively.
It is worth noting that after every two consecutive residual blocks and a batch normalization layer, the max-pooling layer is added. Therefore, there are a total of three max-pooling layers in the model. After pooling layers, dropout is the immediate next layer, which is used in the model to prevent it from over-fitting. It is a regularization technique that randomly drops neurons and their incoming and outgoing edges from a layer, which helps to learn the features in different manners and improves the model's performance.
3.4.4 Time distributed layer
The time-distributed layer is suitable when working with time-series data or video frames. It allows each input to have its own layer. For example, rather than having many "models" for each input, we can have "one model" for each input. Then Bi-GRU can assist with managing the data in "time" and checking the images' previous and future sequences.
3.4.5 Bi-GRU layer
After flattening the features, a bidirectional gated recurrent unit (Bi-GRU), a type of recurrent neural network, is used (RNN). Bi-GRU is a sequential model which calculates the continuity and dependency of features in sequential data, e.g., textual data. Hence, the Bi-GRU layer is used in the proposed model after the feature extraction phase. It computes the dependency and connection between features of the input data [19]. It then passes these features to the fully connected layer in the model based on these characteristics for classification.
Let us divide the input image x into patches M * N \(, { x}_{m, n }\epsilon {R}^{{w}_{p }\times {h}_{p }\times c}\) where, \({w}_{p}\) and\({h}_{p}\), c are the height, width, and the number of channels of the patch respectively and \(M= \frac{w}{{w}_{p}}\), \(N= \frac{h}{{h}_{p}}\), and by inputting \({x}_{m,n}\) in a Bi-GRU layer in a sequence.
Here \({f}_{\mathrm{FWD}}\) and \({f}_{\mathrm{REV}}\) check the dependency of the features of input data in both forward and reverse sequences, respectively, as shown in Eqs. (9) and (10). There is one Bi-GRU layer in the presented model. A hybrid of residual layers and a Bi-GRU layer is used in the model. The Bi-GRU keeps the information about previous and future images, allowing for more accurate forecasting [44].
3.4.6 Flatten and dense layers
The flatten layer reduces the feature map to a single column, which can then be transferred to the next layer for further processing. The dense layer is identified as the fully connected layer in neural networks. In the dense layer, the previous layer's output is converted into a single vector. It determines which feature map best matches a given class by considering the previous layer's output. It gives the correct possibilities for the various classes by using the activation function.
3.4.7 Activation function
ReLU, sigmoid, and softmax activation functions are applied in the proposed model. ReLU is an activation function used in the middle layers of the DNN. In contrast, sigmoid and softmax are used at the output layer for two-class and multi-class classification, respectively.
In Eqs. (11) and (12), x is a number, where \(- n\le x\le n\).
The combination of these layers and activation functions makes our architecture distinctive and novel for detecting non-COVID and COVID-19 pneumonia with different categories of COVID-19. The recommended model is small and lightweight compared to other proposed models described in the discussions section, as fewer parameters are used in this model. It has also performed better than other published studies on a huge dataset.
3.4.8 Discussion on the proposed model
An overview of our proposed Lightweight ResGRU model is described in Fig. 6 and Table 6. According to Fig. 6 and Table 6, the proposed model contains six residual blocks and one Bi-GRU layer. Residual blocks have many advantages over simple CNN architectures. One of the significant advantages is that while increasing the depth of the neural network, it accelerates the training process of the network by using skip connections. The skip connection in the residual block is the backbone that provides direct and short paths from the early layers and connects them 2–3 layers deeper in the neural network, as shown in Fig. 8, which contributes to faster training as well as rapid convergence of the model to higher accuracy. There are two significant purposes of skip connections in the residual block: Firstly, it prevents the vanishing gradient problem; as a result, the network weights and biases will update effectively. Secondly, it overcomes the issue of accuracy saturation in much deeper neural networks [18].
On the other hand, the Bi-GRU layer is used after the feature extraction of the input data. This layer aims to compute the dependencies and connections between input data features [19], improving classification accuracy. It also makes addressing the problem of intra-class similarities (i.e., similarities between images of different classes) easier because it stores the information of the previous and next images of a current image in the data and predicts a class based on this information.
Table 6 contains details of the end-to-end Lightweight ResGRU model, including descriptions of the layers, activations, and learnable weights. The proposed model was trained over 50 epochs with an initial learning rate of 0.001 using the stochastic gradient descent (SGD) optimizer.
4 Experimental setup
This part defines the experimental arrangements used to implement and train the proposed architecture. Experiments are performed on the Google Collaboratory Pro in the Python programming language using the TensorFlow framework. The training process used 25 GB of RAM with a T4 GPU. In the training period of the system, the proposed Lightweight ResGRU was trained end to end through backpropagation using the stochastic gradient descent (SGD) optimizer [45] with an initial learning rate of 0.001, and the batch size during the training phase was set to 8. The input size of images is [224, 224, 3]. The model was trained on 50 epochs, which were appropriate for the convergence of the model. Models with the best validation accuracy were chosen for testing. Furthermore, the model's data splitting and performance evaluation are described in the following sub-sections.
4.1 Dataset splitting
The whole dataset is distributed into three splits, i.e., training, validation, and testing, in the ratio of 80:10:10. Total data samples in each split for different experiments are given in Table 7.
4.2 Performance evaluation
Evaluation measures are applied to the final model to assess the performance of the trained model by using unseen data. The evaluation measures used in our study are confusion matrix, accuracy, precision, sensitivity, specificity, F-measure, false-positive rate (FPR), and false-negative rate (FNR). These evaluation measures are based on true positive (TP), false negative (FN), false positive (FP), and true negative (TN), which are calculated using a confusion matrix. These metrics are described in Eqs. (13) to (19).
TP and TN are correctly predicted as positive and negative cases by the model, respectively. FP and FN show incorrectly predicted positive and negative cases, respectively.
5 Results
This section displays the results and discusses the system's performance on the testing data. In this section, we evaluated the performance of our approach in comparison to previously published studies on the detection of chest pneumonia using four-class classification, three-class classification, and two-class classification.
Table 8 displays the performance of the proposed Lightweight ResGRU in terms of accuracy, precision, sensitivity, specificity, F-measure, FPR, and FNR of four-class classification, three-class classification, two-class classification, and COVID-19 severity classification on a test dataset.
Confusion matrices displayed an outstanding accomplishment of the proposed model on unseen data. In Fig. 9, confusion matrices for four-class, three-class, two-class, and severity classification are plotted. All the results show how accurately our proposed model was trained.
The model accuracy and loss graphs are plotted in Figs. 10 and 11. These graphs illustrate that the proposed model is trained very well as it gives promising results even on unseen data; hence, it is not under-fit or over-fit.
The training graphs show that the model is generalized better on training data. The accuracies and losses for the training and validation sets for the four-class classification are 97.32%, 92%, 0.08, and 0.32, respectively. For three-class classification, the training and validation accuracies are 99.40% and 97.95%, respectively, while the training and validation losses are 0.02 and 0.08, respectively. For multi-modal two-class classification, the training and validation accuracies are 98.42% and 97.61%, respectively, and the training and validation losses are 0.05 and 0.10, respectively. Training and validation accuracies for COVID-19 severity classification are 85.3% and 70.57%, respectively. In comparison, training loss and validation loss for COVID-19 severity classification are 0.40 and 0.9 in the order given.
6 External cohort evaluation: Cross-dataset validation
The final model is evaluated on an external dataset (called cross-dataset validation) to check the robustness of the model. The dataset used for cross-dataset validation is the Curated Dataset of COVID-19 [20]. For cross-dataset validation, randomly selected images are used. The details of the images are provided in Table 9.
Table 10 displays the results of the chosen Lightweight ResGRU for four-class, three-class, and two-class classifications on the external cohort in terms of accuracy, precision, sensitivity, specificity, F-measure, FPR, and FNR. These assessment metrics show how effectively our recommended model has been trained.
Figure 12 shows the confusion matrices for four-class, three-class, and two-class classifications. Confusion matrices revealed that the model performs well on an external dataset as it achieved outstanding results on an external dataset that was not included in training and validation splits.
The encouraging results of the Lightweight ResGRU for identifying chest pneumonia along with the COVID-19 chest infection and its different severity levels in CRIs show that, in the coming time, deep learning models will play a lead role in the clinical diagnosis of this epidemic.
7 Discussions
It can be shown from the results that the proposed model achieved outstanding results on the test set of the development dataset and excellent performance on the external cohort.
Chest X-rays and CT images of patients infected with COVID-19 are the imaging modalities that allow data scientists to work with medical staff. Chest X-rays and CTs are utilized to identify numerous diseases, and CRIs are more extensively used to study brain tumors, heart diseases, and chest infections because they contain more detailed information about the disease. The main signs of the COVID-19 epidemic, which the WHO first reported in late 2019, are severe coughing and breathing difficulties, and X-rays and CT scans are commonly used to diagnose such symptoms. However, there are some bottlenecks and challenges to using imaging-based solutions in which the quantity and quality of data are two concepts that have a direct impact on the deep learning model's performance. Deep learning models need a hefty amount of data for the training to perform better and more successfully. The proposed study generates a large dataset to address these challenges by integrating and modifying different available benchmark datasets.
Our proposed model shows that, by using a hybrid model containing residual blocks and Bi-GRU, a better model can be built for diagnosing chest infections because it predicts by keeping the information of previous and future images in the data. It can be seen from the results that there is an acute sensitivity, which means lower false-negative results. It is worth mentioning that a high false-negative prediction is perilous for society and patients. In that case, the infected people are declared healthy, which is dangerous for healthy people because the infection can spread from these healthy-declared people.
A comparison of various state-of-the-art models reveals that the proposed model has fewer layers and parameters (weights and biases) than other architectures, as shown in Table 11. Hence, the proposed model with just around 6 million parameters is lightweight and has a simple and small architecture, thereby reducing the computational cost of the deep learning model, as presented in Table 6.
Table 11 sums up the studies on the automatic identification of COVID-19 based on chest radiography images and compares them with the proposed model. In Table 11, it can be shown that all the researchers have used deep learning architectures to diagnose chest pneumonia using CRIs. However, most of them used pre-trained models and modified their architectures using transfer learning [46]. Most researchers trained deep learning models on a small dataset, which may result in overfitting and misleadingly good results [36]. Researchers have used a validation dataset instead of a separate unseen test dataset to evaluate deep learning models, leading to biased results and high accuracy scores [37]. On the other hand, the proposed study used an extensive dataset of around forty thousand images for training, validation, testing, and cross-dataset validation of the deep learning model. It assessed the model with unseen datasets (test data and cross-dataset) for unbiased evaluation. The suggested model predicts the four distinct severity levels of COVID-19 (i.e., negative for pneumonia, atypical, indeterminate, and typical), which, to the best of our knowledge, is the first work to conduct a classification of COVID-19 severity levels.
The proposed model is trained and evaluated on a dataset used by another recent study to assess the efficacy of the recommended hybrid architecture. The comparison of the results showed that the proposed architecture achieved better results even when using the same dataset. The model is trained on the SARS-CoV-2 Ct-Scan dataset [23], which Kogilavani et al. [47] used to predict COVID-19 chest infection. The dataset is distributed into three splits, i.e., training, validation, and testing, in the ratio of 70:15:15 after augmentation. Thus, the suggested lightweight model is superior to other models as it performs well on large unseen datasets (test dataset and cross-dataset). It can predict the COVID-19 infection and its severity level. Lightweight ResGRU achieved an overall testing accuracy of 98.56% and a sensitivity of 97.17%. Table 12 shows the results of Lightweight ResGRU compared with the study [47], where the same dataset was used in both cases.
8 Conclusion and future works
The proposed study used a novel and hybrid model to diagnose non-COVID and COVID-19 chest infections, including their different severity types, using chest radiography images (X-rays and CTs). It is a hybrid model based on multiple residual blocks and Bi-GRU that detects clinical features and the dependency of those features, respectively. As a result, it provides promising classification results. The presented deep learning model named Lightweight ResGRU is proficient in the diagnosis of 2 class multi-modal (X-rays, CTs) classification (normal and COVID-19), 3 class classification (normal, COVID-19, and viral pneumonia), 4 class classification (normal, COVID-19, viral pneumonia, and bacterial pneumonia). It can also classify different severity types (atypical, indeterminate, typical, and negative for pneumonia) of COVID-19, which, to the best of our knowledge, has not been published yet. The proposed model achieved 99.0%, 98.4%, 91.0%, and 80.5% f-measure and 0.009, 0.08, 0.02, and 0.19 FNR for two-class, three-class, four-class classification, and COVID-19 severity classification respectively, on an extensive test dataset.
Another contribution of the study is the acquisition and creation of a large dataset for chest infection prediction. The performance of the proposed Lightweight ResGRU shows the power of this method over the existing research studies. The future aim is to overcome resource restrictions, making us capable of training the model on a larger dataset. We expect to further increase the model's performance, especially for the external cohort, by using a significantly high number of images in the training phase. Another objective is to use the object detection algorithm on CRIs to localize the infected region of the lungs, which can provide a better understanding of the patient’s condition to the radiologists.
Data availability
The links to the datasets created and analyzed during the current study can be found in the [Lightweight ResGRU] repository, [https://github.com/Mughees-Ahmad/Lightweight_ResGRU].
References
Huang C et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395(10223):497–506. https://doi.org/10.1016/S0140-6736(20)30183-5
Wu F et al (2020) A new coronavirus associated with human respiratory disease in China. Nature 579(7798):265–269. https://doi.org/10.1038/s41586-020-2008-3
“Coronavirus (COVID-19) events as they happen.” https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen (accessed Dec. 02, 2021).
“WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard With Vaccination Data.” https://covid19.who.int/ (accessed Dec. 02, 2021).
Bhatt T, Kumar V, Pande S, Malik R, Khamparia A, Gupta D (2021) A Review on COVID-19. Stud Comput Intell 924(April):25–42. https://doi.org/10.1007/978-3-030-60188-1_2
Hussain E, Hasan M, Rahman MA, Lee I, Tamanna T, Parvez MZ (2021) “CoroDet: A deep learning based classification for COVID-19 detection using chest X-ray images”, Chaos. Solitons and Fractals 142:110495. https://doi.org/10.1016/j.chaos.2020.110495
Das AK, Kalam S, Kumar C, Sinha D (2021) “TLCoV- An automated Covid-19 screening model using transfer learning from chest X-ray images”, Chaos. Solitons and Fractals 144:541. https://doi.org/10.1016/j.chaos.2021.110713
Lee EYP, Ng MY, Khong PL (2020) COVID-19 pneumonia: what has CT taught us? Lancet Infect Dis 20(4):384–385. https://doi.org/10.1016/S1473-3099(20)30134-1
Benmalek E, Elmhamdi J, Jilbab A (2021) Comparing CT scan and chest X-ray imaging for COVID-19 diagnosis. Biomed Eng Adv 1:100003. https://doi.org/10.1016/j.bea.2021.100003
Naguib M, Moustafa F, Salman MT, Saeed NK, Al-Qahtani M (2020) The use of radiological imaging alongside reverse transcriptase PCR in diagnosing novel coronavirus disease 2019: a narrative review. Future Microbiol 15(10):897–903. https://doi.org/10.2217/fmb-2020-0098
Chan JFW et al (2020) A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395(10223):514–523. https://doi.org/10.1016/S0140-6736(20)30154-9
Kong W, Agarwal PP (2020) Chest imaging appearance of covid-19 infection. Radiol Cardiothorac Imaging 2(1):2–5. https://doi.org/10.1148/ryct.2020200028
Prokop M et al (2020) CO-RADS: a categorical CT assessment scheme for patients suspected of having COVID-19-definition and evaluation. Radiology 296(2):E97–E104. https://doi.org/10.1148/radiol.2020201473
de Jaegere TMH, Krdzalic J, Fasen BACM, Kwee RM (2020) Radiological society of north america chest ct classification system for reporting covid-19 pneumonia: Interobserver variability and correlation with reverse-transcription polymerase chain reaction. Radiol Cardiothorac Imaging 2:3. https://doi.org/10.1148/ryct.2020200213
Yang S, Gao T, Wang J, Deng B, Lansdell B, Linares-Barranco B (2021) Efficient spike-driven learning with dendritic event-based processing. Front Neurosci 15:55. https://doi.org/10.3389/fnins.2021.601109
Holzinger A, Langs G, Denk H, Zatloukal K, Müller H (2019) “Causability and explainability of artificial intelligence in medicine”, Wiley Interdiscip. Rev Data Min Knowl Discov 9(4):1–13. https://doi.org/10.1002/widm.1312
Krizhevsky BA, Sutskever I, Hinton GE (2012) Cnn实际训练的. Commun ACM 60(6):84–90
He K, Zhang X, Ren S, Sun J (2016) “Deep residual learning for image recognition. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2015:770–778. https://doi.org/10.1109/CVPR.2016.90
Yin Q, Zhang R, Shao X (2019) CNN and RNN mixed model for image classification. MATEC Web Conf 277:02001. https://doi.org/10.1051/matecconf/201927702001
U. SAIT et al., “Curated Dataset for COVID-19 Posterior-Anterior Chest Radiography Images (X-Rays).,” vol. 1, 2020, doi: https://doi.org/10.17632/9XKHGTS2S6.1.
“COVID-19 Radiography Database | Kaggle.” https://www.kaggle.com/tawsifurrahman/covid19-radiography-database (accessed May 19, 2021).
“SIIM-FISABIO-RSNA COVID-19 Detection | Kaggle.” https://www.kaggle.com/c/siim-covid19-detection (accessed Dec. 06, 2021).
“SARS-COV-2 Ct-Scan Dataset | Kaggle.” https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset (accessed May 19, 2021).
Ardakani AA, Kanafi AR, Acharya UR, Khadem N, Mohammadi A (2020) Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 121:103795. https://doi.org/10.1016/j.compbiomed.2020.103795
Shibly KH, Dey SK, Islam MTU, Rahman MM (2020) COVID faster R-CNN: a novel framework to diagnose novel coronavirus disease (COVID-19) in X-Ray images. Informatics Med Unlocked 20:100405. https://doi.org/10.1016/j.imu.2020.100405
Javor D, Kaplan H, Kaplan A, Puchner SB, Krestan C, Baltzer P (2020) Deep learning analysis provides accurate COVID-19 diagnosis on chest computed tomography. Eur J Radiol 133:109402. https://doi.org/10.1016/j.ejrad.2020.109402
Wang S et al (2021) A deep learning algorithm using CT images to screen for Corona virus disease (COVID-19). Eur Radiol 31(8):6096–6104. https://doi.org/10.1007/s00330-021-07715-1
Ismael AM, Şengür A (2020) Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst Appl 164(September):2021. https://doi.org/10.1016/j.eswa.2020.114054
Dash AK, Mohapatra P (2021) A Fine-tuned deep convolutional neural network for chest radiography image classification on COVID-19 cases. Multimed Tools Appl 15:1. https://doi.org/10.1007/s11042-021-11388-9
Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Rajendra Acharya U (2020) Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med 121:10379. https://doi.org/10.1016/j.compbiomed.2020.103792
Islam MZ, Islam MM, Asraf A (2020) A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Informatics Med. Unlocked 20:100412. https://doi.org/10.1016/j.imu.2020.100412
Demir F (2021) DeepCoroNet: A deep LSTM approach for automated detection of COVID-19 cases from chest X-ray images. Appl Soft Comput 103:107160. https://doi.org/10.1016/j.asoc.2021.107160
Turkoglu M (2021) COVIDetectioNet: COVID-19 diagnosis system based on X-ray images using features selected from pre-learned deep features ensemble. Appl Intell 51(3):1213–1226. https://doi.org/10.1007/s10489-020-01888-w
Luz E et al (2022) Towards an effective and efficient deep learning model for COVID-19 patterns detection in X-ray images. Res Biomed Eng 38(1):149–162. https://doi.org/10.1007/s42600-021-00151-6
Elkorany AS, Elsharkawy ZF (2021) COVIDetection-Net: a tailored COVID-19 detection from chest radiography images using deep learning. Optik (Stuttg) 231:166405. https://doi.org/10.1016/j.ijleo.2021.166405
“Why Aren’t My Results As Good As I Thought? You’re Probably Overfitting.” https://machinelearningmastery.com/arent-results-good-thought-youre-probably-overfitting/ (accessed May 19, 2022).
“What is the Difference Between Test and Validation Datasets?” https://machinelearningmastery.com/difference-test-validation-datasets/ (accessed May 19, 2022).
Kleppe A, Skrede OJ, De Raedt S, Liestøl K, Kerr DJ, Danielsen HE (2021) Designing deep learning studies in cancer diagnostics. Nat Rev Cancer 21(3):199–211. https://doi.org/10.1038/s41568-020-00327-9
M. de la I. Vayá et al., “BIMCV COVID-19+: a large annotated dataset of RX and CT images from COVID-19 patients,” pp. 1–22, 2020, [Online]. Available: http://arxiv.org/abs/2006.01174
“Medical Imaging Data Resource Center (MIDRC) - RSNA International COVID-19 Open Radiology Database (RICORD) Release 1c - Chest x-ray Covid+ (MIDRC-RICORD-1c) - The Cancer Imaging Archive (TCIA) Public Access - Cancer Imaging Archive Wiki.” https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=70230281 (accessed May 19, 2021).
Swati ZNK et al (2019) Brain tumor classification for MR images using transfer learning and fine-tuning. Comput Med Imaging Graph 75:34–46. https://doi.org/10.1016/j.compmedimag.2019.05.001
Lecun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444. https://doi.org/10.1038/nature14539
S. Ioffe and C. Szegedy, “Batch normalization: Accelerating deep network training by reducing internal covariate shift,” 32nd Int. Conf. Mach. Learn. ICML 2015, 1: 448–456, 2015.
D. M. Ibrahim, N. M. Elshennawy, and A. M. Sarhan, “Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID- 19. The COVID-19 resource centre is hosted on Elsevier Connect, the company ’ s public news and information,” no. January, 2020.
Liu Y, Gao Y, Yin W (2020) An improved analysis of stochastic gradient descent with momentum. Adv Neural Inf Process Syst 2:1–11
Weiss K, Khoshgoftaar TM, and Wang DD, A survey of transfer learning, vol. 3, no. 1. Springer International Publishing, 2016. doi: https://doi.org/10.1186/s40537-016-0043-6.
Kogilavani SV et al. (2022) “COVID-19 Detection Based on Lung Ct Scan Using Deep Learning Techniques,” Comput Math Methods Med, vol. 2022,. doi: https://doi.org/10.1155/2022/7672196.
El Asnaoui K, Chawki Y (2020) Using X-ray images and deep learning for automated detection of coronavirus disease. J Biomol Struct Dyn 39(10):1–12. https://doi.org/10.1080/07391102.2020.1767212
Gilanie G et al (2021) Coronavirus (COVID-19) detection from chest radiology images using convolutional neural networks. Biomed Signal Process Control 6:102490. https://doi.org/10.1016/j.bspc.2021.102490
Kumar A, Tripathi AR, Satapathy SC, Zhang YD (2022) SARS-Net: COVID-19 detection from chest x-rays by combining graph convolutional network and convolutional neural network. Pattern Recognit 122:108255. https://doi.org/10.1016/j.patcog.2021.108255
Funding
The authors have no relevant financial or non-financial interests to disclose.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors have no competing interests to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ahmad, M., Bajwa, U.I., Mehmood, Y. et al. Lightweight ResGRU: a deep learning-based prediction of SARS-CoV-2 (COVID-19) and its severity classification using multimodal chest radiography images. Neural Comput & Applic 35, 9637–9655 (2023). https://doi.org/10.1007/s00521-023-08200-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-023-08200-0