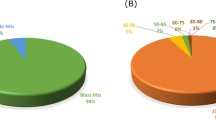
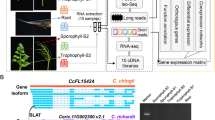
Abstract
Apogamy is a phenomenon in which a sporophyte develops asexually, directly from a cell or cells of a gametophyte. It is a phenomenon described mainly in lower plants, but shares certain aspects with apomixis in angiosperms. The genes involved in apogamy commitment in ferns are unknown. We hypothesize that the mechanism of asexual reproduction is controlled in lower and higher plants by overlapping sets of genes. To this end, we created a normalized subtracted cDNA library that represents genes with increased expression during apogamy commitment in the fern Ceratopteris richardii. The cDNA library consists of 170 unique sequences. Compared to the mature gametophyte transcriptome of the fern Pteridium aquilinum, the apogamy library is enriched in plant GO-Slim terms that are associated with stress and metabolism. In silico expression analyses of the closest Arabidopsis homologs of the apogamy library revealed many genes that display preferential expression in seed and flower tissues, structures that are absent in ferns. This apogamy library provides a rich resource for investigations into the genetic control of apogamy in ferns and comparisons with the asexual processes of higher plants.






Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarkis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet 25:25–29
Asker SE, Jerling L (1992) Apomixis in plants. CRC Press, Boca Raton
Banks JA (1994) Sex-determining genes in the homosporous fern Ceratopteris. Development 120:1949–1958
Banks JA (1997) Sex determination in the fern Ceratopteris. Trends Plant Sci 2:175–180
Byrne ME (2009) A role for the ribosome in development. Trends Plant Sci 14:512–519
Chaudhury AM, Ming L, Miller C, Craig S, Dennis ES, Peacock WJ (1997) Fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 94:4223–4228
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Ann Biochem 162:156–159
Conesa A, Gotz S (2008) Blast2GO: a comprehensive suite for functional analysis in plants genomics. Int J Plant Genomics 2008:1–13
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Cordle AR, Irish EE, Cheng CL (2007) Apogamy induction in Ceratopteris richardii. Int J Plant Sci 168:361–369
Cordle AR, Bui LT, Cheng CL (2010) Laboratory-induced apogamy and apospory in Ceratopteris richardii. In: Fernandez H, Kumar A, Revilla MA (eds) Working With Ferns: Issues and Applications, 1st edn. Springer, New York, pp 25–36
Der JP, Barker MS, Wickett NJ, dePamphilis CW, Wolf PG (2011) De novo characterization of the gametophyte transcriptome in bracken fern, Pteridium aquilinum. BMC Genomics 12:99–112
DeYoung B, Weber T, Hass T, Banks JA (1997) Generating autotetraploid sporophytes and their use in analyzing mutations affecting gametophyte development in the fern Ceratopteris. Genetics 147:809–814
Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA 93:6025–6030
Farlow WG (1874) An asexual growth from the prothallus of Pteris cretica. Q J Microsc Sci 14:266–272
Hecht V, Vielle-Calzada JP, Hartog MV, Schmidt EDL, Boutillier K, Grossniklaus U, deVries SC (2001) The Arabidopsis Somatic Embryogenesis Receptor Kinase 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol 127:803–816
Hennig L, Derkacheva M (2009) Diversity of polycomb group complexes in plants: same rules, different players? Trends Genet 25:414–423
Hey SJ, Byrne E, Halford NG (2009) The interface between metabolic and stress signaling. Ann Bot 105:197–203
Holec S, Berger F (2012) Polycomb group complexes mediate developmental transitions in plants. Plant Physiol 158:35–43
Jackson S, Rounsley S, Purugganan M (2006) Comparative sequencing of plant genomes: choices to make. Plant Cell 18:1100–1104
Klekowski EJ (1979) The genetics and reproductive biology of ferns. In: Dyer AF (ed) The experimental biology of ferns. Academic Press, New York, pp 133–165
Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chadhury AM (1999) Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96:96–301
Martinoia E, Maeshima M, Neuhaus HE (2006) Vacuolar transporters and their essential role in plant metabolism. J Exp Bot 58:83–102
Mosquna A, Katz A, Decker EL, Rensing SA, Reski R, Ohad N (2009) Regulation of stem cell maintenance by the polycomb protein FIE has been conserved during land plant evolution. Development 136:2433–2444
Okano Y, Aono N, Hiwatashi Y, Murata T, Nishiyama T, Ishkawa T, Kubo M, Hasebe M (2009) A polycomb repressive complex 2 gene regulates apogamy and gives evolutionary insights into early land plant evolution. Proc Natl Acad Sci USA 106:16321–16326
Pires ND, Dolan L (2012) Morphological evolution in land plants: new designs with old genes. Philos Trans R Soc Lond Ser B Biol Sci 367:508–518
Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–W120
Raghavan V (1989) Developmental biology of fern gametophytes. Cambridge University Press, Cambridge
Salmi ML, Bushart TJ, Stout SC, Roux S (2005) Profile and analysis of gene expression changes during early development in germinating spores of Ceratopteris richardii. Plant Physiol 138:1734–1745
Schmidt EDL, Guzzo F, Toonen MAJ, de Vries SC (1997) A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 124:2049–2062
Segui-Simarro JM (2010) Androgenesis revisited. Bot Rev 76:377–404
Shariatpanahi ME, Bal U, Heberle-Bors E, Touraev A (2006) Stresses applied for the reprogramming of plant microspores towards in vitro embryogenesis. Physiol Plant 127:519–534
Sheffield E, Bell PR (1987) Current studies of the pteridophyte life cycle. Bot Rev 53:442–490
Steeves TA, Sussex IM, Partanen CR (1955) In vitro studies on abnormal growth of prothalli of the bracken fern. Am J Bot 42:232–245
Tucker MR, Koltunow MG (2009) Sexual and asexual (apomictic) seed development in flowering plants: molecular, morphological and evolutionary relationships. Funct Plant Biol 36:490–504
Walker TG (1979) The cytogenetics of ferns. In: Dyer AF (ed) The experimental biology of ferns. Academic press, New York, pp 87–123
Whittier DP (1964) The influence of cultural conditions on the induction of apogamy in Pteridium gametophytes. Am J Bot 51:730–736
Whittier DP (1966) The influence of growth substances on the induction of apogamy in Pteridium gametophytes. Am J Bot 53:882–886
Whittier DP (1975) The influence of osmotic conditions on induced apogamy in Pteridium gametophytes. Phytomorphology 25:246–249
Whittier DP, Pratt LH (1971) The effect of light quality on the induction of apogamy in prothalli of Pteridium aquilinum. Planta 99:174–178
Whittier DP, Steeves TA (1960) The induction of apogamy in the bracken fern. Can J Bot 38:925–930
Whittier DP, Steeves TA (1962) Further studies on induced apogamy in ferns. Can J Bot 40:1525–1531
Zimmerman P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2622
Acknowledgments
We would like to thank J. Fassler (University of Iowa) for critical readings of the manuscript, and D. Houston (University of Iowa) and G. Hehman (Carver Center for Genomics) for assistance with the RT-qPCR experiments and sequencing. This work was supported by a grant from the University of Iowa Research Council. ARC was also supported by Avis Cone Summer Fellowships.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Scott Russell.
Electronic supplementary material
Below is the link to the electronic supplementary material.
497_2012_198_MOESM2_ESM.xls
The identity, lengths, minimum blast E-values, Arabidopsis homologues, and Plant GO-Slim terms mapped to the 170 apogamy library sequences 2 (XLS 128 kb)
Rights and permissions
About this article
Cite this article
Cordle, A.R., Irish, E.E. & Cheng, CL. Gene expression associated with apogamy commitment in Ceratopteris richardii . Sex Plant Reprod 25, 293–304 (2012). https://doi.org/10.1007/s00497-012-0198-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00497-012-0198-z