Abstract
Key message
A novel cold responsive HbICE1 gene from Hevea brasiliensis was identified, and its function as a transcriptional factor in cold stress responses was further analyzed.
Abstract
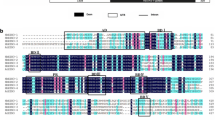
The inducers of CBF expression (ICEs) play a crucial role in the response of plants to cold stress. To date, information is limited about the ICE genes from rubber trees (Hevea brasiliensis Muell. Arg.), a tropical woody plant. In this study, an ICE homologuous gene, referred to as HbICE1, was isolated and characterized by a combination of molecular and physiological techniques and bioinformatics analyses. The full-length cDNA of HbICE1 was 1806-bp in length and contained a 1617 bp open reading frame (ORF) encoding a putative protein of 538 amino acids. The predicted HbICE1 harbored S-rich, bHLH and ACT-like domains that were conserved in ICE1 orthologs from Arabidopsis and other plant species. Phylogenetic analysis demonstrated that HbICE1 was closely related to MeICE1 and RcICE1 as well as PeICE1, MdICE1 and PuICE1. It harbored transcriptional activity and nucleus localization. qRT-PCR analysis showed that HbICE1 was mainly expressed in leaves and stems under natural conditions. Its expression was significantly up-regulated in the leaves of the cold-resistant rubber tree clone ‘93-114’ in response to cold stress by comparison with the cold-sensitive rubber tree clone ‘Reken501’. Functional analysis in yeast revealed that HbICE1 conferred cold stress resistance. Collectively, these results suggest that HbICE1 acts as a transcriptional factor and indicatively mediates the cold resistance in rubber trees.






Similar content being viewed by others
Abbreviations
- CBF:
-
C-repeat binding transcription factors
- df:
-
Degree of freedom
- GFP:
-
Green fluorescence protein
- ICE:
-
Inducer of CBF expression
- tBLASTn:
-
Translated basic local alignment search tool
- qRT-PCR:
-
Real-time quantitative reverse transcription PCR
- bHLH:
-
Basic helix-loop-helix
- ZIP:
-
Zipper region
- RT-PCR:
-
Reverse transcription PCR
- pI:
-
Isoelectric point
- SC:
-
Synthetic complete medium
- SE:
-
Standard deviation
References
Badawi M, Reddy YV, Agharbaoui Z, Tominaga Y, Danyluk J, Sarhan F, Houde M (2008) Structure and functional analysis of wheat ICE (inducer of CBF expression) genes. Plant Cell Physiol 49:1237–1249
Budhagatapalli N, Narasimhan R, Rajaraman J, Viswanathan C, Nataraja KN (2015) Ectopic expression of AtICE1and OsICE1 transcription factor delays stress-induced senescence and improves tolerance to abiotic stresses in tobacco. J Plant Biochem Biotechnol. doi:10.1007/s13562-015-0340-8
Chao J, Zhang S, Chen Y, Tian WM (2015) Cloning, heterologous expression and characterization of ascorbate peroxidase (APX) gene in laticifer cells of rubber tree (Hevea brasiliensis Muell. Arg.). Plant Physiol Biochem 97:331–338
Chen L, Lopes JM (2010) Multiple bHLH proteins regulate CIT2 expression in Saccharomyces cerevisiae. Yeast 27:345–359
Chen L, Chen Y, Jiang J, Chen S, Chen F, Guan Z, Fang W (2012) The constitutive expression of Chrysanthemum dichrum ICE1 in Chrysanthemum grandiflorum improves the level of low temperature, salinity and drought tolerance. Plant Cell Rep 31:1747–1758
Chen Y, Jiang J, Song A, Chen S, Shan H, Luo H, Gu C, Sun J, Zhu L, Fang W, Chen F (2013) Ambient temperature enhanced freezing tolerance of Chrysanthemum dichrum CdICE1 Arabidopsis via miR398. BMC Biol 11:121
Cheng H, Cai H, Fu H, An Z, Fang J, Hu Y, Guo D, Huang H (2015) Functional Characterization of Hevea brasiliensis CRT/DRE Binding Factor 1 Gene Revealed Regulation Potential in the CBF Pathway of Tropical Perennial Tree. PLoS ONE 10:e0137634
Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17:1043–1054
Chinnusamy V, Zhu J, Zhu JK (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12:444–451
Chinnusamy V, Zhu JK, Sunkar R (2010) Gene regulation during cold stress acclimation in plants. Methods Mol Biol 639:39–55
Deng X, Hu W, Wei S, Zhou S, Zhang F, Han J, Chen L, Li Y, Feng J, Fang B, Luo Q, Li S, Liu Y, Yang G, He G (2013) TaCIPK29, a CBL-interacting protein kinase gene from wheat, confers salt stress tolerance in transgenic tobacco. PLoS ONE 8:e69881
Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S (2015) OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev Cell 32(3):278–289
Duan C, Rio M, Leclercq J, Bonnot F, Oliver G, Montoro P (2010) Gene expression pattern in response to wounding, methyl jasmonate and ethylene in the bark of Hevea brasiliensis. Tree Physiol 30:1349–1359
Feller A, Hernandez JM, Grotewold, E (2006) An ACT-like Domain Participates in the dimerization of several plant basic-helix-loop-helix transcription factors. J Biol Chem 281(39):28964–28974
Feng XM, Zhao Q, Zhao LL, Qiao Y, Xie XB, Li HF, Yao YX, You CX, Hao YJ (2012) The cold-induced basic helix-loop-helix transcription factor gene MdCIbHLH1 encodes an ICE-like protein in apple. BMC Plant Biol 12:22
Feng HL, Ma NN, Meng X, Zhang S, Wang JR, Chai S, Meng QW (2013) A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant Physiol Biochem 73:309–320
Gujjar RS, Akhtar M, Singh M (2014) Transcription factors in abiotic stress tolerance. Ind J Plant Physiol 19:306–316
Hu Y, Jiang L, Wang F, Yu D (2013) Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25:2907–2924
Huang GT, Ma SL, Bai LP, Zhang L, Ma H, Jia P, Liu J, Zhong M, Guo ZF (2012) Signal transduction during cold, salt, and drought stresses in plants. Mol Biol Rep 39:969–987
Huang X, Li K, Jin C, Zhang S (2015a) ICE1 of Pyrus ussuriensis functions in cold tolerance by enhancing PuDREB a transcriptional levels through interacting with PuHHP1. Sci Rep 5:17620
Huang XS, Zhang Q, Zhu D, Fu X, Wang M, Zhang Q, Moriguchi T (2015b) Liu JH (2015b) ICE1 of Poncirus trifoliata functions in cold tolerance by modulating polyamine levels through interacting with arginine decarboxylase. J Exp Bot 66:3259–3274
Jiao Z, Bedoui S, Brady JL, Walter A, Chopin M, Carrington EM, Sutherland RM, Nutt SL, Zhang Y, Ko HJ, Wu L, Lew AM, Zhan Y (2014) The closely related CD103+ dendritic cells (DCs) and lymphoid-resident CD8+ DCs differ in their inflammatory functions. PLoS ONE 9:e91126
Kong Q, Pattanaik S, Feller A, Werkman JR, Chai C, Wang Y, Grotewold E, Yuan L (2012) Regulatory switch enforced by basic helix-loop-helix and ACT-domain mediated dimerizations of the maize transcription factor R. Proc Natl Acad Sci USA 109:E2091–E2097
Lee BH, Henderson DA, Zhu JK (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant cell 17(11):3155–3175
Lei X, Xiao Y, Xia W, Mason AS, Yang Y, Ma Z, Peng M (2014) RNA-seq analysis of oil palm under cold stress reveals a different C-repeat binding factor (CBF) mediated gene expression pattern in Elaeis guineensis compared to other species. PLoS ONE 9:e114482
Li H, Qin Y, Xiao X, Tang C (2011) Screening of valid reference genes for real-time RT-PCR data normalization in Hevea brasiliensis and expression validation of a sucrose transporter gene HbSUT3. Plant Sci 181:132–139
Li X, Zhang D, Li H, Wang Y, Zhang Y, Wood AJ (2014) EsDREB2B, a novel truncated DREB2-type transcription factor in the desert legume Eremosparton songoricum, enhances tolerance to multiple abiotic stresses in yeast and transgenic tobacco. BMC Plant Biol 14:44
Lin YZ, Zheng HQ, Zhang Q, Liu CX, Zhang ZY (2013) Functional profiling of EcaICE1 transcription factor gene from Eucalyptus camaldulensis involved in cold response in tobacco plants. J Plant Biochem Biotechnol 23:141–150
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time Quantitative PCR and the 2–ΔΔCt method. Methods 25:402–408
Peng HH, Shan W, Kuang JF, Lu WJ, Chen JY (2013) Molecular characterization of cold-responsive basic helix-loop-helix transcription factors MabHLHs that interact with MaICE1 in banana fruit. Planta 238:937–953
Peng PH, Lin CH, Tsai HW, Lin TY (2014) Cold response in Phalaenopsis aphrodite and characterization of PaCBF1 and PaICE1. Plant Cell Physiol 55:1623–1635
Rahman AY, Usharraj AO, Misra BB, Thottathil GP, Jayasekaran K, Feng Y, Hou S, Ong SY, Ng FL, Lee LS, Tan HS, Sakaff MK, The BS, Khoo BF, Badai SS, Aziz NA, Yuryev A, Knudsen B, Dionne-Laporte A, Mchunu NP, Yu Q, Langston BJ, Freitas TA, Young AG, Chen R, Wang L, Najimudin N, Saito JA, Alam M (2013) Draft genome sequence of the rubber tree Hevea brasiliensis. BMC Genom 14:75
Robinson KA, Lopes JM (2000) SURVEY AND SUMMARY: Saccharomyces cerevisiae basic helix-loop-helix proteins regulate diverse biological processes. Nucleic Acids Res 28:1499–1505
Schade B, Jansen G, Whiteway M, Entian KD, Thomas DY (2004) Cold adaptation in budding yeast. Mol Biol Cell 15:5492–5502
Shan W, Kuang JF, Lu WJ, Chen JY (2014) Banana fruit NAC transcription factor MaNAC1 is a direct target of MaICE1 and involved in cold stress through interacting with MaCBF1. Plant, Cell Environ 37:2116–2127
Shi Y, Ding Y, Yang S (2015) Cold signal transduction and its interplay with phytohormones during cold acclimation. Plant Cell Physiol 56(1):7–15
Silva CC, Mantello CC, Campos T, Souza LM, Gonçalves PS, Souza AP (2014) Leaf-, panel- and latex-expressed sequenced tags from the rubber tree (Hevea brasiliensis) under cold-stressed and suboptimal growing conditions: the development of gene-targeted functional markers for stress response. Mol Breed 34:1035–1053
Sun H, Fan HJ, Ling HQ (2015) Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genom 16:9
Teste MA, Duquenne M, François JM, Parrou JL (2009) Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol Biol 10:99
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034
Wang X, Sun X, Liu S, Liu L, Liu X, Sun X, Tang K (2005) Molecular cloning and characterization of a novel ice gene from Capsella bursapastoris. Mol Biol 39:21–29
Wang Y, Jiang CJ, Li YY, Wei CL, Deng WW (2012) CsICE1 and CsCBF1: two transcription factors involved in cold responses in Camellia sinensis. Plant Cell Rep 31:27–34
Xiang DJ, Hu XY, Zhang Y, Yin KD (2008) Over-Expression of ICE1 gene in transgenic rice improves cold tolerance. Rice Sci 15:173–178
Xu W, Jiao Y, Li R, Zhang N, Xiao D, Ding X, Wang Z (2014) Chinese wild-growing Vitis amurensis ICE1 and ICE2 encode MYC-type bHLH transcription activators that regulate cold tolerance in Arabidopsis. PLoS ONE 9:e102303
Zhan X, Zhu JK, Lang Z (2015) Increasing freezing tolerance: kinase regulation of ICE1. Dev Cell 32(3):257–258
Zhou LH, Wu JL, Chen S, Juan L (2012) Cold-induced modulation of CbICE53gene activates endogenous genes to enhance acclimation in transgenic tobacco. Mol Breeding 30:1611–1620
Acknowledgments
This work was supported by the earmarked fund for China Agriculture Research System (CARS-34-GW1) and the Fundamental Research Funds for Rubber Research Institute, CATAS (1630022014002). We thank National Infrastructure for Crop Germplasm Resources-National Infrastructure for Rubber Tree Germplasm Resources for providing budwoods of rubber tree clone ‘93-114’ and ‘Reken501’.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The paper has no conflict of interest.
Additional information
Communicated by J. Carlson.
Xiao Min Deng and Jian Xiao Wang have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Deng, X.M., Wang, J., Li, Y. et al. Characterization of a cold responsive HbICE1 gene from rubber trees. Trees 31, 137–147 (2017). https://doi.org/10.1007/s00468-016-1463-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-016-1463-9