Abstract
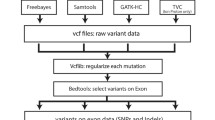
We assessed the performance of the new Life Technologies Proton sequencer by comparing whole-exome sequence data in a Centre d’Etude du Polymorphisme Humain trio (family 1463) to the Illumina HiSeq instrument. To simulate a typical user’s results, we utilized the standard capture, alignment and variant calling methods specific to each platform. We restricted data analysis to include the capture region common to both methods. The Proton produced high quality data at a comparable average depth and read length, and the Ion Reporter variant caller identified 96 % of single nucleotide polymorphisms (SNPs) detected by the HiSeq and GATK pipeline. However, only 40 % of small insertion and deletion variants (indels) were identified by both methods. Usage of the trio structure and segregation of platform-specific alleles supported this result. Further comparison of the trio data with Complete Genomics sequence data and Illumina SNP microarray genotypes documented high concordance and accurate SNP genotyping of both Proton and Illumina platforms. However, our study underscored the problem of accurate detection of indels for both the Proton and HiSeq platforms.


Similar content being viewed by others
References
Bras J, Guerreiro R, Hardy J (2012) Use of next-generation sequencing and other whole-genome strategies to dissect neurological disease. Nat Rev Neurosci 13:453–464
Cancer Genome Atlas N (2012) Comprehensive molecular portraits of human breast tumours. Nature 490:61–70
Clark MJ, Chen R, Lam HY, Karczewski KJ, Chen R, Euskirchen G, Butte AJ, Snyder M (2011) Performance comparison of exome DNA sequencing technologies. Nat Biotechnol 29:908–914
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST et al (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
Dausset J, Cann H, Cohen D, Lathrop M, Lalouel JM, White R (1990) Centre d’etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics 6:575–577
Dean M (2003) Approaches to identify genes for complex human diseases: lessons from Mendelian disorders. Hum Mutat 22:261–274
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M et al (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498
Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G et al (2010) Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327:78–81
Gilissen C, Hoischen A, Brunner HG, Veltman JA (2012) Disease gene identification strategies for exome sequencing. Eur J Hum Genet: EJHG 20:490–497
Gonzaga-Jauregui C, Lupski JR, Gibbs RA (2012) Human genome sequencing in health and disease. Annu Rev Med 63:35–61
Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L et al (2011) Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet 43:875–878
Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA Jr, Vogelstein B et al (2010) Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330:228–231
Lam HY, Clark MJ, Chen R, Natsoulis G, O’Huallachain M, Dewey FE, Habegger L, Ashley EA, Gerstein MB, Butte AJ et al (2012) Performance comparison of whole-genome sequencing platforms. Nat Biotechnol 30:78–82
Liu P, Morrison C, Wang L, Xiong D, Vedell P, Cui P, Hua X, Ding F, Lu Y, James M et al (2012) Identification of somatic mutations in non-small cell lung carcinomas using whole-exome sequencing. Carcinogenesis 33:1270–1276
Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ (2012) Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 30:434–439
Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA et al (2010) Whole-genome sequencing in a patient with Charcot–Marie-Tooth neuropathy. N Engl J Med 362:1181–1191
Manolio TA, Chisholm RL, Ozenberger B, Roden DM, Williams MS, Wilson R, Bick D, Bottinger EP, Brilliant MH, Eng C et al (2013) Implementing genomic medicine in the clinic: the future is here. Genet Med 15(4):258–267
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Merriman B, IT RDT, Rothberg JM (2012) Progress in Ion Torrent semiconductor chip based sequencing. Electrophoresis 33:3397–3417
Meyerson M, Gabriel S, Getz G (2010) Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 11:685–696
Nickles D, Madireddy L, Yang S, Khankhanian P, Lincoln S, Hauser SL, Oksenberg JR, Baranzini SE (2012) In depth comparison of an individual’s DNA and its lymphoblastoid cell line using whole genome sequencing. BMC Genomics 13:477
O’Rawe J, Jiang T, Sun G, Wu Y, Wang W, Hu J, Bodily P, Tian L, Hakonarson H, Johnson WE et al (2013) Low concordance of multiple variant-calling pipelines: practical implications for exome and genome sequencing. Genome Med 5:28
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575
Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y (2012) A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13:341
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842
Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M et al (2011) An integrated semiconductor device enabling non-optical genome sequencing. Nature 475:348–352
St Hilaire C, Ziegler SG, Markello TC, Brusco A, Groden C, Gill F, Carlson-Donohoe H, Lederman RJ, Chen MY, Yang D et al (2011) NT5E mutations and arterial calcifications. N Engl J Med 364:432–442
Veltman JA, Brunner HG (2012) De novo mutations in human genetic disease. Nat Rev Genet 13:565–575
Wang Z, Jacobs KB, Yeager M, Hutchinson A, Sampson J, Chatterjee N, Albanes D, Berndt SI, Chung CC, Diver WR et al (2012) Improved imputation of common and uncommon SNPs with a new reference set. Nat Genet 44:6–7
Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N (2012) LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 40:11189–11201
Acknowledgments
This research has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E and supported in part by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and the Center for Cancer Research, National Cancer Institute, NIH, DHHS, Bethesda, MD, USA. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services or does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Boland, J.F., Chung, C.C., Roberson, D. et al. The new sequencer on the block: comparison of Life Technology’s Proton sequencer to an Illumina HiSeq for whole-exome sequencing. Hum Genet 132, 1153–1163 (2013). https://doi.org/10.1007/s00439-013-1321-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-013-1321-4