Abstract
Genetic variation on the Y chromosome has not been convincingly implicated in prostate cancer risk. To comprehensively analyze the role of inherited Y chromosome variation in prostate cancer risk in individuals of European ancestry, we genotyped 34 binary Y chromosome markers in 3,995 prostate cancer cases and 3,815 control subjects drawn from four studies. In this set, we identified nominally significant association between a rare haplogroup, E1b1b1c, and prostate cancer in stage I (P = 0.012, OR = 0.51; 95% confidence interval 0.30–0.87). Population substructure of E1b1b1c carriers suggested Ashkenazi Jewish ancestry, prompting a replication phase in individuals of both European and Ashkenazi Jewish ancestry. The association was not significant for prostate cancer overall in studies of either Ashkenazi Jewish (1,686 cases and 1,597 control subjects) or European (686 cases and 734 control subjects) ancestry (P meta = 0.078), but a meta-analysis of stage I and II studies revealed a nominally significant association with prostate cancer risk (P meta = 0.010, OR = 0.77; 95% confidence interval 0.62–0.94). Comparing haplogroup frequencies between studies, we noted strong similarities between those conducted in the US and France, in which the majority of men carried R1 haplogroups, resembling Northwestern European populations. On the other hand, Finns had a remarkably different haplogroup distribution with a preponderance of N1c and I1 haplogroups. In summary, our results suggest that inherited Y chromosome variation plays a limited role in prostate cancer etiology in European populations but warrant follow-up in additional large and well characterized studies of multiple ethnic backgrounds.
Similar content being viewed by others
Introduction
Family and twin studies have shown that prostate cancer has a clear heritable component which may be among the highest of all cancer types (Amundadottir et al. 2004; Lichtenstein et al. 2000), Over the last few years, genome wide association studies (GWAS) have successfully identified germline variants conferring risks of prostate cancer at over 45 loci (Amundadottir et al. 2006; Chung and Chanock 2011; Eeles et al. 2008, 2009; Gudmundsson et al. 2007a, b, 2008, 2009; Haiman et al. 2007; Kote-Jarai et al. 2011; Schumacher et al. 2011; Takata et al. 2010; Thomas et al. 2008; Yeager et al. 2007, 2009). These studies have not implicated variants on the Y chromosome in the risk of prostate cancer, possibly due to the fact that very few Y chromosome SNPs have been included on most genotyping chips used to date. Several groups have specifically investigated the role of Y chromosome haplogroups in prostate cancer risk. Many of these studies are inconclusive due to the small number of samples and/or markers used. One of the larger studies was conducted within the multi-ethnic cohort (MEC) using samples from prostate cancer cases and control subjects drawn from four ethnic groups. Of the 41 haplogroups observed, one was significantly associated with prostate cancer in Japanese men (Paracchini et al. 2003) but this association was not replicated in a separate study from Korea (Kim et al. 2007). No association was seen between Y haplogroups and prostate cancer in a large Swedish study (Lindstrom et al. 2008).
The Y chromosome contains the largest non-recombining region in the human genome, spanning almost the entire length of the chromosome. This region is called the non-recombining Y (NRY) or the male-specific Y (MSY) (Rozen et al. 2003). In the absence of recombination, the NRY passes mostly unchanged from father to son and observed mutations reflect the evolutionary history of the Y chromosome. Binary markers can be used to classify Y chromosomes into haplogroups organized by a phylogenetic tree. A first generation phylogeny of the tree was published in 2002 by the Y Chromosome Consortium (2002) and further revised in 2008 (Karafet et al. 2008). The Y chromosome tree now consists of over 300 haplogroups organized into 20 major groups or clades (Karafet et al. 2008).
Multiple lines of evidence support a possible role for genes on the Y chromosome in prostate cancer etiology. Loss of the Y chromosome is one of the most frequent cytogenetic change seen in prostate tumors and may be an early event in tumorigenesis (Brothman et al. 1999; Jordan et al. 2001). In support of the previous assertion, chromosome transfer studies indicate that the human Y chromosome suppresses tumorigenicity of human prostate cell lines in vivo implying that it may harbor gene(s) with tumor suppressor function (Vijayakumar et al. 2005). Based on the essential role of the Y chromosome in secondary sexual differentiation and its potential role in disease pathogenesis, particularly related to the secondary sex organs, we explored this genomic region to investigate whether germline variation on this chromosome plays a role in prostate cancer risk.
Results
We analyzed 7,810 men from the Cancer Genetic Markers of Susceptibility (CGEMS) scan in stage I of this study. Of the 34 chromosome Y markers genotyped, 26 were observed in our sample (8 markers were monomorphic). With such a sample size, we were able to accurately characterize and estimate the Y chromosome frequency distribution in populations of European ancestry for 28 haplogroups including three combined groups (R1b1b + R1b*, R1a + R1* and I2b + I2c) as the leaf nodes of the NRY tree (Fig. 1a). Stage I had 41, 76 and 95% power to detect an association with an odds ratio of 1.3 and a MAF of 0.02, 0.05 and 0.10, respectively (assuming prostate cancer prevalence of 1.5067% and alpha of 0.05) (http://seer.cancer.gov/csr/1975_2007/).
Chromosome Y haplogroup tree and frequency distribution in control subjects of European ancestry in Stage I. a Chromosome Y tree showing genotyped markers in black and those not genotyped in light grey. Haplogroup names are according to the International Society of Genetic Genealogy (ISOGG) 2011 update. The arrow points to the mutational event which gave rise to the E1b1b1c haplogroup. Stage I studies are the following: CPS-II American Cancer Society Cancer Prevention Study II, ATBC Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study, CeRePP Centre de Recherche pour les Pathologies Prostatiques, and PLCO Prostate, Lung Colorectal and Ovarian Cancer Screening Trial. b The circle plots show frequencies for haplogroups with a derived frequency of 5% or higher in different colors for each Stage I cohort (remaining haplogroups are combined in one group shown in black)
Stage I association analysis
After genotyping quality control based on completion rates and concordance analysis, a total of 3,995 prostate cancer cases and 3,815 control subjects from four studies were used in the analysis (1994; Calle et al. 2002; Gohagan et al. 2000; Valeri et al. 2003). This included 1,531 men diagnosed with non-aggressive prostate cancer (Gleason score <7 and disease stage <III) and 2,142 men diagnosed with aggressive prostate cancer (Gleason score ≥7 or stage ≥III).
Of the 26 haplogroup markers analyzed, one was significantly associated with overall prostate cancer at a nominal P value threshold of P ≤ 0.05 (Table 1). This was haplogroup E1b1b1c (locus M123, formerly named haplogroup E3b1c) (P = 0.012, allelic odds ratio (OR) 0.51; 95% confidence interval 0.30–0.87), a rare haplogroup with a 1.1% frequency in control subjects in our sample set. When the analysis was performed according to degree of differentiation and severity of prostate cancer, this haplogroup was significantly associated with non-aggressive prostate cancer (P = 0.017, allelic OR 0.33; 95% confidence interval 0.13–0.86) but not with aggressive prostate cancer (P = 0.091, allelic OR 0.59; 95% confidence interval 0.32–1.09). However, the difference between the two case groups was not significant (P = 0.48).
Based on the Y chromosome haplogroup tree structure (see “Methods” section and Fig. 1a), we were able to test three additional haplgroups (J1, IJ, IJK) for which markers were not directly genotyped. These three haplogroups did not significantly associate with overall risk of prostate cancer (data not shown) (Table 1).
Population substructure of E1b1b1c carriers
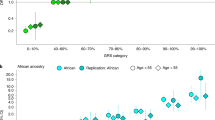
We assessed the population substructure for E1b1b1c haplogroup carriers using principal components analysis (PCA) from the initial CGEMS GWAS dataset (Thomas et al. 2008; Yeager et al. 2007, 2009). In addition we evaluated one common haplogroup, R1b1a2 (43.7% frequency in control subjects from stage I) as it had the second lowest P value in stage I (P = 0.054). Carriers of the E1b1b1c haplogroup showed a distinct distribution of the first and second eigenvectors (EV1 and EV2) in this analysis that separates them from the majority of the European ancestry subjects, i.e. negative values for EV1 and positive values for EV2 (Fig. 2a). Conversely, the population substructure of R1b1a2 haplogroup carriers was similar to that of the majority of subjects in our study, implying Northwestern European ancestry. We compared the population substructure pattern of E1b1b1c haplogroup carriers to the large number of individuals in the initial CGEMS prostate cancer scan (Thomas et al. 2008; Yeager et al. 2007, 2009) and a GWAS of breast cancer in families of Ashkenazi Jewish descent (Gold et al. 2008) and noted strong clustering with a group of individuals of self-reported Ashkenazi Jewish descent with similar values for EV1 and EV2 (Fig. 2b). The majority of E1b1b1c haplogroup carriers in our study (37 out of 62) were Ashkenazi Jewish alike by this comparison. Therefore, the frequency of E1b1b1c in inferred Ashkenazi Jewish individuals in our study was estimated to be approximately 15% (37/240). A similar number has been reported in men of Jewish ancestry (Hammer et al. 2009). These findings prompted a replication phase in sample sets of both European and Ashkenazi Jewish ancestry.
Population substructure analysis by principal component analysis and comparison to CGEMS prostate cancer GWAS. a shows the distribution of the first two principal components, EV1 and EV2, for carriers of E1b1b1c (filled squares) and R1b1a2 (open circles) haplogroups in Stage I. Circles and squares denote eigenvalues from PCA analysis for each individual. The distribution of EV1 and EV2 for all Stage I subjects is shown in b. Studies are designed by different colors. CPS-II Blood blood derived DNA samples were used for genotyping, CPS-II Buccal buccal derived DNA samples were used for genotyping. DNA samples from ATBC, CeRePP and PLCO were all derived from blood. Individuals of inferred Ashkenazi Jewish ancestry are circled. PCA results were performed by EIGENSTRAT in CGEMS prostate cancer GWAS (Thomas et al. 2008; Yeager et al. 2007, 2009)
Limited evidence for association to prostate cancer in Stage II analysis
We attempted replication of the E1b1b1c and R1b1a2 haplogroups in three prostate cancer cohort studies of European ancestry from the continental USA, the Physicians’ Health Study (PHS) (Ma et al. 2008), the Health Professionals Follow-up Study (HPFS) (Chen et al. 2005), the Agricultural Health Study (AHS) (Alavanja et al. 1996); and in two case–control studies of Ashkenazi Jewish ancestry collected in the USA, from the Albert Einstein College of Medicine (Einstein) (Agalliu et al. 2009) and the Memorial Sloan Kettering Cancer Center (MSKCC) (Gallagher et al. 2010). The three European ancestry studies included a total of 1,272 prostate cancer cases and 1,932 control subjects; the two Ashkenazi Jewish ancestry studies included a total of 1,686 prostate cancer cases and 1,597 control subjects. Neither haplogroup was significantly associated with overall prostate cancer risk at a nominal P value in any study (Table 2) nor was a meta-analysis of the combined studies significant (P meta = 0.078 for E1b1b1c and P meta = 0.36 for R1b1a2). For non-aggressive prostate cancer, the E1b1b1c haplogroup was significantly associated at a nominal P value in the Einstein study only (P = 0.024, allelic OR 0.66; 95% confidence interval 0.45–0.95). However, a meta-analysis of non-aggressive cases in all stage II studies was nominally significant (P meta = 0.025, allelic OR 0.68; 95% confidence interval 0.49–0.95). No other significant associations were noted.
A meta-analysis of stage I and II results for the E1b1b1c haplogroup revealed a nominally significant association with risk of prostate cancer overall (P meta = 0.010; allelic OR = 0.77; 95% confidence interval 0.62–0.94) and with risk of non-aggressive prostate cancer (P meta = 0.0077; allelic OR = 0.67; 95% confidence interval 0.50–0.90) but not with risk of aggressive prostate cancer (P meta = 0.28). In Stage II, we had 60% power to detect a variant with 13% MAF and an OR of 0.8 in the Ashkenazi Jewish sample set, but only 25% power to detect a variant with 3% MAF and an OR of 0.7 in the European American sample set.
Haplogroup frequency and population distribution
Y chromosome haplogroup frequency distribution in controls from each of the four study populations from phase I was summarized and compared in Fig. 1b. Two of the studies, namely CPS-II and PLCO, include subjects from continental USA. Their haplogroup frequencies are very similar with an average difference of 0.8% and a maximum difference of 5.8% for the combined category of haplogroups R1b1b + R1b*. The CeRePP study, conducted in France, is relatively similar to the US studies with an average haplogroup frequency difference of 2.2%, and a maximum difference of 24.3% for the combined group of R1b1b + R1b*. The greatest difference in frequency was seen for ATBC, a Finnish study, with an average haplogroup frequency difference of 5.2% and a maximum difference of 54.4%. This stems from a very high frequency of haplogroup N1c in this study (55.6%), while it is infrequent in the other three studies from the US and France (0.8% in CPS-II, 1.7% in PLCO and 0.4% in CeRePP). Second, R1b, the most frequent haplogroup overall, is seen in over 50% of subjects in PLCO, CPS-II and CeRePP but only in 4.8% of Finnish subjects. The third largest difference was noted for haplogroup I1 which was more common in Finns at 27.6%, as compared to 13.9% in PLCO, 11.2% in CPS-II and only 8.1% in CeRePP.
Haplogroup E1b was observed at low frequencies in all studies and its sub lineage E1b1b1c was seen in approximately 1–2% of subjects from the two US studies (PLCO and CPS-II) and the French study (CeRePP), whereas it was absent from the Finnish study (ATBC). Other haplogroups were absent or rare in the four studies.
Discussion
In this study, we explored the role of germline Y chromosome variation in prostate cancer risk. Previous studies have not analyzed such a large sample size with as many markers in individuals of European ancestry. Because of the threshold for MAF chosen for this study (≥1%), we had limited capacity to detect risk variants with low to medium frequency and effect sizes. Prostate cancer GWAS to date have used arrays with limited coverage on the Y chromosome. As an example, in CGEMS, of the approximately 500,000 SNPs genotyped in stage I, only ten Y chromosome markers passed quality control assessment and were included in the primary analysis; this limited set of variants on the Y chromosome included only four that mark chromosome Y haplogroups (Thomas et al. 2008; Yeager et al. 2007, 2009). Other published prostate cancer GWAS studies have reported on a similar fraction of Y variants (Amundadottir et al. 2006; Chung and Chanock 2011; Eeles et al. 2008, 2009; Gudmundsson et al. 2007a, b, 2008, 2009; Haiman et al. 2007; Kote-Jarai et al. 2011; Schumacher et al. 2011; Takata et al. 2010; Thomas et al. 2008; Yeager et al. 2007, 2009).
One haplogroup of interest was noted in phase I of our study; the E1b1b1c haplogroup was nominally significant in the overall prostate cancer and non-aggressive prostate cancer groups. The marker that denotes this haplogroup is located in the last intron of the taxilin gamma 2 pseudogene (TXLNG2P) on chromosome Yq11.222. This haplogroup was analyzed in a second phase using replication studies of European and Ashkenazi Jewish ancestry along with a more common haplogroup, R1b1a2. Neither haplogroup was significantly associated with overall prostate cancer risk in stage II. A meta-analysis of stage I and stage II results yielded a P value of 0.010 for the E1b1b1c haplogroup. Although nominally significant, this P value is unremarkable in comparison with the rigorous threshold required for significance in GWAS studies (Wellcome Trust Case Control Consortium 2007), suggesting that further studies are required to establish this association. Although our analysis does not provide strong evidence for a relationship between variation in the Y chromosome and prostate cancer, it can be argued that the appropriate statistical threshold to be applied to a study of approximately 30 markers should not be as stringent as a GWAS threshold. However, the probability of false-positive findings is high, even in a study of our size and power (Wacholder et al. 2004) especially in the first stage where E1b1b1c haplogroup frequency was very low. In addition, we cannot exclude a chance finding due to population stratification.
Our study represents the largest analysis to date of a possible association between Y chromosome variants and prostate cancer. The role of germline variation on the Y chromosome had been assessed previously, but with limited sample and/or marker sets. One of the most complete studies published was conducted within the MEC (Paracchini et al. 2003). Four ethnic groups with a total of 930 cases and 1,208 control subjects were included. One of the 41 haplogroups observed in the study was significantly associated with prostate cancer risk in Japanese men with a P value of 0.02 (Paracchini et al. 2003). Despite the large overall sample set in this study, each ethnic group only consisted of approximately 100–150 case–control pairs, limiting power considerably. No haplogroups were significantly associated with prostate cancer risk in a small Korean study that assessed 14 markers in approximately 106 cases and 110 control subjects, including the haplogroup reported in the MEC study (Kim et al. 2007). Lack of an association between Y haplogroups and prostate cancer was also reported in a Swedish study assessing five ChrY markers in 1,452 cases and 779 control subjects of N-European background (Lindstrom et al. 2008). Our results appear to confirm an overall lack of importance for germline variants on the Y chromosome and prostate cancer risk.
Frequencies of Y chromosome haplogroups vary considerably between different geographical regions and ethnic groups, and have turned out to be informative in studies of human evolution and migration. In Europe, marked differences in haplogroup frequencies are observed between countries in Northeastern, Northwestern, Southwestern, Southeast and Central Europe (Wiik 2008). In addition, the Ashkenazi Jewish community has a specific pattern that is reminiscent of non-Ashkenazi Jewish communities in the Near East (Behar et al. 2004). We observed a different distribution of major haplogroups in subjects of Northwestern European ancestry (represented by the majority of subjects from the US in PLCO and CPS-II), Northeastern European ancestry (represented by Finnish subjects in ATBC) and Western/Central European ancestry (represented by French subjects in CeRePP). Haplogroups in the US and French studies can mostly be accounted for by the R and I haplogroup clans with a combined frequency of 81–85%; R1b1a2 and I1 were the most common sub branches. The R1 haplogroup clan originated in Eurasia and migrated into Europe where it divided into two subgroups, R1a (common in Eastern Europe) and R1b (common in Western Europe) (Wiik 2008). R1b1a2 shows an East to West gradient in Europe and is very common in Spain, France, UK and Ireland (Balaresque et al. 2010). Haplogroup clan I1 appears to have originated in the Balkans and migrated north throughout Europe (Wiik 2008). It is most common in Scandinavia and Northwestern Europe and gradually decreases in Central and Southern Europe (Wiik 2008). Finnish subjects were strikingly different from the other three studies with a preponderance of N1c (56%) and I1 (28%) haplogroups and few R1b carriers. The N1c haplogroup is thought to have an Eastern or Central Asian origin and probably reached Eastern Europe via expansion through Siberia (Rootsi et al. 2007). The frequency of this haplogroup in Finland has been reported to be 58% (Wiik 2008).
Genotypes in stage II confirmed the scarcity of E1b1b1c in subjects of European ancestry (1–2%) and revealed a higher frequency in the two Ashkenazi Jewish studies (13–14%), in line with previous reports (Hammer et al. 2009) indicating similar Y chromosome haplogroup frequencies in men of Ashkenazi Jewish descent living in the US and those from Jewish communities in the Middle East. E1b1b1c may have arisen in Northeastern Africa, and migrated through the Levantine corridor to the Near East and Europe (Semino et al. 2004). In a similar manner, haplogroup R1b1a2 was seen in 50–59% of the subjects in different European American studies but only 10–11% in the two Ashkenazi Jewish studies.
In conclusion, we found limited evidence for an association between Y chromosome haplogroups and risk of prostate cancer in populations of European and Ashkenazi Jewish ancestry using a large sample set close to 4,000 case–control pairs in Stage I and 2,300 case–control pairs in Stage II. Weak but consistent evidence for a protective effect for haplogroup E1b1b1c was seen in all studies with a nominally significant meta-analysis, thus, calling for additional replication efforts for this haplogroup in populations of Ashkenazi Jewish and European ancestry. The different frequencies seen in subjects from the four stage I studies may limit power to detect true associations for some branches of the Y haplogroup tree. Furthermore, correcting for population substructure based on autosomal SNPs may not be optimal, as Y chromosome inheritance only reflects male lineages that may have somewhat different characteristics throughout human history and population migration as compared to that of females. Although we cannot exclude a role for all chromosome Y haplogroups in prostate cancer etiology, our study has good power to detect common alleles with relatively large effects. Smaller or population specific effects for the haplgroups tested here, or for other haplogroups, could exist and should be studied by testing comprehensive sets of chromosome Y haplogroup markers in additional studies.
Materials and methods
Study population
Stage I of this study included 3,995 men diagnosed with adenocarcinoma of the prostate and 3,815 control subjects from three case–control studies nested within cohorts and one hospital based case–control study, previously analyzed in stages I and II of the Cancer Genetics Markers of Susceptibility study (CGEMS). Study details have been published previously (Thomas et al. 2008; Yeager et al. 2007, 2009).The cohort studies were: the Prostate, Lung Colorectal and Ovarian Cancer Screening Trial (PLCO, subjects from continental USA) (Gohagan et al. 2000); the American Cancer Society Cancer Prevention Study II (CPS-II, from continental USA) (Calle et al. 2002) and the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC, from Finland) (1994). The case–control study was the French Prostate Case–Control Study (CeRePP, Centre de Recherche pour les Pathologies Prostatiques, from France) (Valeri et al. 2003). The number of subjects included from each study is shown in Supplemental Table 1a. We incorporated prostate cancer stage and grade at diagnosis to distinguish between non-aggressive (Gleason score <7 and disease stage <III, n = 1,531) and aggressive prostate cancer (Gleason score ≥7 and/or disease stage ≥III, n = 2,141) as defined in CGEMS (Thomas et al. 2008).
Stage II included 471 prostate cancer cases and 490 control subjects of European descent from the Physicians’ Health Study (PHS, from continental USA) (Ma et al. 2008); 215 prostate cancer cases and 244 control subjects of European descent from the Health Professionals Follow-up Study (HPFS, from continental USA) (Chen et al. 2005); 586 prostate cancer cases and 1198 control subjects of European descent from the Agricultural Health Study (AHS, from NC and IA) (Alavanja et al. 1996); 933 prostate cancer cases and 1,221 control subjects of Ashkenazic descent collected by the Albert Einstein College of Medicine (Einstein, majority recruited from NY, FL, CA or NJ, USA) (Agalliu et al. 2009); and 753 prostate cancer cases and 376 male control subjects of Ashkenazic descent collected at the Memorial Sloan Kettering Cancer Center (MSKCC, from Northeast USA) (Gallagher et al. 2010). Prostate cancer stage and grade at diagnosis were included to distinguish between non-aggressive (Gleason score <7 AND disease stage <III, n = 194 for PHS; n = 172 for HPFS, n = 338 for AHS; n = 416 for Einstein and n = 212 for MSKCC) and aggressive prostate cancer (Gleason score ≥7 OR disease stage ≥III, n = 167 for PHS; n = 80 for HPFS, n = 85 for AHS; n = 457 for Einstein and n = 364 for MSKCC).
The study protocols for each study were approved by the Institutional Review Board of each corresponding institution, and written informed consent was obtained from all study participants.
Marker selection and genotyping
Markers were selected to detect chromosome Y haplogroups with minor allele frequencies (MAF) ≥1% in populations of European descent, using data from the International Society of Genetic Genealogy (ISOGG) (http://www.isogg.org/tree/ISOGG_YDNA_SNP_Index.html) 2011 update, the Y Chromosome Consortium (http://ycc.biosci.arizona.edu/) (Karafet et al. 2008; Underhill et al. 2001) and from HapMap (http://hapmap.ncbi.nlm.nih.gov/). TaqMan custom genotyping assays (ABI, Foster City, CA, USA) were designed and optimized for 34 biallelic chromosome Y markers (32 SNPs and 2 insertion/deletion polymorphisms) based on the Y Chromosome Consortium, ISOGG and HapMap databases.
For stage I, DNA was extracted from blood samples for all studies except a subset of CPS-II where buccal cells were used for a subset of subjects (n = 939). After pre-genotyping quality control at the Core Genotyping Facility (CGF) of the National Cancer Institute of the National Institutes of Health, Gaithersburg, MD, USA (http://cgf.nci.nih.gov/operations/pregenotyping-qaqc.html), 34 SNPs were genotyped on 9,501 samples in stage I using TaqMan genotyping assays (ABI, Foster City, CA, USA). The average concordance for 146 duplicate samples was 99.75%. Samples were excluded based on a completion rate <80% or ≥2 heterozygous genotypes. After genotype quality control (Supplemental Table 1a), 8,157 samples remained (including 8,011 subjects of which 7,810 men (3,995 cases and 3,815 controls) had all covariates used in the association analysis). Eight markers were monomorphic in our data set (Supplemental Table 2), thus leaving 26 polymorphic markers for analysis.
For stage II, DNA was isolated from blood for all studies except for the samples from Einstein where DNA was obtained from mouthwash. A subset of DNA samples from the AHS study (n = 1,858) were whole genome amplified prior to genotyping using the GenomiPhi™ version 2 kit (GE Healthcare) at the Core Genotyping Facility (CGF) of the National Cancer Institute of the National Institutes of Health, Gaithersburg, MD, USA (http://cgf.nci.nih.gov/operations/wga.html). Two SNPs (M123 for haplogroup E1b1b1c and M269 for haplogroup R1b1a2) were genotyped in stage II on 6,695 samples using TaqMan genotyping assays (ABI, Foster City, CA, USA) at CGF and on 1,213 samples at MSKCC (for MSKCC samples). This included 6,487 subjects (2,958 case and 3,529 control subjects). The E1b1b1c haplogroup was genotyped in samples from Einstein, MSKCC, PHS and HPFS; the R1b1a2 haplogroup was genotyped in samples from Einstein, MSKCC, PHS and AHS. Genotype quality control was performed in a similar manner as for stage I studies (detailed in Supplemental Table 1b). Concordance rates for duplicate samples (n = 91) were 99.9%.
Statistical analysis
The association between haplogroups of the Y chromosome and prostate cancer risk was examined using a logistic regression model adjusted for age, study center and first principle component previously constructed based on CGEMS genotype data (Thomas et al. 2008; Yeager et al. 2007, 2009) as it was significant in the base model, to correct for population stratification if available. All subjects were of self-described European ancestry.
The variance weighted fixed-effect meta-analysis was performed to assess the overall statistical significance of stage II studies as well as combination of stage I together with II studies. Results were not corrected for multiple testing because of the strong dependence among the markers on this chromosome. Because all the Y haplogroups map to a haplogroup evolutionary tree, each branch in the tree can be cut and thus creating a bipartition of all individuals. Individuals under each cut will have inherited the mutation incurred on that branch. The case/control imbalance could therefore be tested by comparing two groupings of subjects. This is exactly the same as testing the genotypes of individual markers. To make full use of the data, genotypes from untyped branches were imputed if possible, based on its ancestor and sibling nodes in the tree. As an example, we could infer genotypes for J1 because both J and J2 were genotyped (Fig. 1a). We searched across all the branches in the tree and tested three additional untyped haplogroups, namely J1 (M267), IJ (M429) and IJK (M522). A branch was not analyzed if there was a directly genotyped derived/ancestor branch with a difference in frequency of <0.001. For example, M96 and M203 are almost the same because frequencies of haplogroup D in all study populations were close to 0. Thus, testing of the imputed marker M203 became redundant when the directly genotyped marker M96 was already analyzed.
Validation by sequencing
Genotypes for the two markers selected for replication (M123 for haplogroup E1b1b1c and M269 for haplogroup R1b1a2) were confirmed by sequencing in 94 subjects from the current study. They were chosen from the PLCO, CeRePP and CPS-II studies in stage I such that approximately one-third (E1b1b1c) or half (R1b1a2) carried each haplogroup. Primers were designed with the program Primer3 (http://frodo.wi.mit.edu/primer3/) and used for PCR amplification of the genomic regions containing the 2 markers (Supplemental Table 3). PCR amplifications were performed with 10 ng genomic DNA using the AmpliTaq Gold 360 master mix (ABI). The samples were cleaned using AMPure beads (Agencourt) on a Biomek FX (Beckman Coulter). After resuspending the beads in 50 μl of water, PCR products were sequenced using primers for the two markers and an ABI PRISM Big Dye Terminator version 3.1 cycle sequencing kit (Applied BioSystems, Foster City, CA, USA). Sequencing was performed on an ABI 3730 capillary sequencer (Applied Biosystems). A 100% concordance was noted for E1b1b1c and 98.9% concordance for R1b1a2.
References
Agalliu I, Gern R, Leanza S, Burk RD (2009) Associations of high-grade prostate cancer with BRCA1 and BRCA2 founder mutations. Clin Cancer Res 15:1112–1120. doi:10.1158/1078-0432.CCR-08-1822
Alavanja MC, Sandler DP, McMaster SB, Zahm SH, McDonnell CJ, Lynch CF, Pennybacker M, Rothman N, Dosemeci M, Bond AE, Blair A (1996) The Agricultural Health Study. Environ Health Perspect 104:362–369
Amundadottir LT, Thorvaldsson S, Gudbjartsson DF, Sulem P, Kristjansson K, Arnason S, Gulcher JR, Bjornsson J, Kong A, Thorsteinsdottir U, Stefansson K (2004) Cancer as a complex phenotype: pattern of cancer distribution within and beyond the nuclear family. PLoS Med 1:e65. doi:10.1371/journal.pmed.0010065
Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, Jakobsdottir M, Kostic J, Magnusdottir DN, Ghosh S, Agnarsson K, Birgisdottir B, Le Roux L, Olafsdottir A, Blondal T, Andresdottir M, Gretarsdottir OS, Bergthorsson JT, Gudbjartsson D, Gylfason A, Thorleifsson G, Manolescu A, Kristjansson K, Geirsson G, Isaksson H, Douglas J, Johansson JE, Balter K, Wiklund F, Montie JE, Yu X, Suarez BK, Ober C, Cooney KA, Gronberg H, Catalona WJ, Einarsson GV, Barkardottir RB, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K (2006) A common variant associated with prostate cancer in European and African populations. Nat Genet 38:652–658. doi:10.1038/ng1808
Balaresque P, Bowden GR, Adams SM, Leung HY, King TE, Rosser ZH, Goodwin J, Moisan JP, Richard C, Millward A, Demaine AG, Barbujani G, Previdere C, Wilson IJ, Tyler-Smith C, Jobling MA (2010) A predominantly neolithic origin for European paternal lineages. PLoS Biol 8:e1000285. doi:10.1371/journal.pbio.1000285
Behar DM, Garrigan D, Kaplan ME, Mobasher Z, Rosengarten D, Karafet TM, Quintana-Murci L, Ostrer H, Skorecki K, Hammer MF (2004) Contrasting patterns of Y chromosome variation in Ashkenazi Jewish and host non-Jewish European populations. Hum Genet 114:354–365. doi:10.1007/s00439-003-1073-7
Brothman AR, Maxwell TM, Cui J, Deubler DA, Zhu XL (1999) Chromosomal clues to the development of prostate tumors. Prostate 38:303–312. doi:10.1002/(SICI)1097-0045(19990301)38:4<303:AID-PROS6>3.0.CO;2-E
Calle EE, Rodriguez C, Jacobs EJ, Almon ML, Chao A, McCullough ML, Feigelson HS, Thun MJ (2002) The American Cancer Society Cancer Prevention Study II Nutrition Cohort: rationale, study design, and baseline characteristics. Cancer 94:2490–2501. doi:10.1002/cncr.101970
Chen YC, Giovannucci E, Lazarus R, Kraft P, Ketkar S, Hunter DJ (2005) Sequence variants of toll-like receptor 4 and susceptibility to prostate cancer. Cancer Res 65:11771–11778. doi:10.1158/0008-5472.CAN-05-2078
Chung CC, Chanock SJ (2011) Current status of genome-wide association studies in cancer. Hum Genet 130:59–78. doi:10.1007/s00439-011-1030-9
Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, Field HI, Southey MC, Severi G, Donovan JL, Hamdy FC, Dearnaley DP, Muir KR, Smith C, Bagnato M, Ardern-Jones AT, Hall AL, O’Brien LT, Gehr-Swain BN, Wilkinson RA, Cox A, Lewis S, Brown PM, Jhavar SG, Tymrakiewicz M, Lophatananon A, Bryant SL, Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Fisher C, Jamieson C, Cooper CS, English DR, Hopper JL, Neal DE, Easton DF (2008) Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet 40:316–321. doi:10.1038/ng.90
Eeles RA, Kote-Jarai Z, Al Olama AA, Giles GG, Guy M, Severi G, Muir K, Hopper JL, Henderson BE, Haiman CA, Schleutker J, Hamdy FC, Neal DE, Donovan JL, Stanford JL, Ostrander EA, Ingles SA, John EM, Thibodeau SN, Schaid D, Park JY, Spurdle A, Clements J, Dickinson JL, Maier C, Vogel W, Dork T, Rebbeck TR, Cooney KA, Cannon-Albright L, Chappuis PO, Hutter P, Zeegers M, Kaneva R, Zhang HW, Lu YJ, Foulkes WD, English DR, Leongamornlert DA, Tymrakiewicz M, Morrison J, Ardern-Jones AT, Hall AL, O’Brien LT, Wilkinson RA, Saunders EJ, Page EC, Sawyer EJ, Edwards SM, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper CS, Southey MC, Lophatananon A, Liu JF, Kolonel LN, Le Marchand L, Wahlfors T, Tammela TL, Auvinen A, Lewis SJ, Cox A, FitzGerald LM, Koopmeiners JS, Karyadi DM, Kwon EM, Stern MC, Corral R, Joshi AD, Shahabi A, McDonnell SK, Sellers TA, Pow-Sang J, Chambers S, Aitken J, Gardiner RA, Batra J, Kedda MA, Lose F, Polanowski A, Patterson B, Serth J, Meyer A, Luedeke M, Stefflova K, Ray AM, Lange EM, Farnham J, Khan H, Slavov C, Mitkova A, Cao G et al (2009) Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet 41:1116–1121. doi:10.1038/ng.450
Gallagher DJ, Vijai J, Cronin AM, Bhatia J, Vickers AJ, Gaudet MM, Fine S, Reuter V, Scher HI, Hallden C, Dutra-Clarke A, Klein RJ, Scardino PT, Eastham JA, Lilja H, Kirchhoff T, Offit K (2010) Susceptibility loci associated with prostate cancer progression and mortality. Clin Cancer Res 16:2819–2832. doi:10.1158/1078-0432.CCR-10-0028
Gohagan JK, Prorok PC, Hayes RB, Kramer BS (2000) The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial of the National Cancer Institute: history, organization, and status. Control Clin Trials 21:251S–272S
Gold B, Kirchhoff T, Stefanov S, Lautenberger J, Viale A, Garber J, Friedman E, Narod S, Olshen AB, Gregersen P, Kosarin K, Olsh A, Bergeron J, Ellis NA, Klein RJ, Clark AG, Norton L, Dean M, Boyd J, Offit K (2008) Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci USA 105:4340–4345. doi:10.1073/pnas.0800441105
Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Xu J, Blondal T, Kostic J, Sun J, Ghosh S, Stacey SN, Mouy M, Saemundsdottir J, Backman VM, Kristjansson K, Tres A, Partin AW, Albers-Akkers MT, Godino-Ivan Marcos J, Walsh PC, Swinkels DW, Navarrete S, Isaacs SD, Aben KK, Graif T, Cashy J, Ruiz-Echarri M, Wiley KE, Suarez BK, Witjes JA, Frigge M, Ober C, Jonsson E, Einarsson GV, Mayordomo JI, Kiemeney LA, Isaacs WB, Catalona WJ, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K (2007a) Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet 39:631–637. doi:ng1999[pii]10.1038/ng1999
Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, Rafnar T, Gudbjartsson D, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Blondal T, Stacey SN, Helgason A, Gunnarsdottir S, Olafsdottir A, Kristinsson KT, Birgisdottir B, Ghosh S, Thorlacius S, Magnusdottir D, Stefansdottir G, Kristjansson K, Bagger Y, Wilensky RL, Reilly MP, Morris AD, Kimber CH, Adeyemo A, Chen Y, Zhou J, So WY, Tong PC, Ng MC, Hansen T, Andersen G, Borch-Johnsen K, Jorgensen T, Tres A, Fuertes F, Ruiz-Echarri M, Asin L, Saez B, van Boven E, Klaver S, Swinkels DW, Aben KK, Graif T, Cashy J, Suarez BK, van Vierssen Trip O, Frigge ML, Ober C, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Palmer CN, Rotimi C, Chan JC, Pedersen O, Sigurdsson G, Benediktsson R, Jonsson E, Einarsson GV, Mayordomo JI, Catalona WJ, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K (2007b) Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet 39: 977–983. doi:10.1038/ng2062
Gudmundsson J, Sulem P, Rafnar T, Bergthorsson JT, Manolescu A, Gudbjartsson D, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Blondal T, Jakobsdottir M, Stacey SN, Kostic J, Kristinsson KT, Birgisdottir B, Ghosh S, Magnusdottir DN, Thorlacius S, Thorleifsson G, Zheng SL, Sun J, Chang BL, Elmore JB, Breyer JP, McReynolds KM, Bradley KM, Yaspan BL, Wiklund F, Stattin P, Lindstrom S, Adami HO, McDonnell SK, Schaid DJ, Cunningham JM, Wang L, Cerhan JR, St Sauver JL, Isaacs SD, Wiley KE, Partin AW, Walsh PC, Polo S, Ruiz-Echarri M, Navarrete S, Fuertes F, Saez B, Godino J, Weijerman PC, Swinkels DW, Aben KK, Witjes JA, Suarez BK, Helfand BT, Frigge ML, Kristjansson K, Ober C, Jonsson E, Einarsson GV, Xu J, Gronberg H, Smith JR, Thibodeau SN, Isaacs WB, Catalona WJ, Mayordomo JI, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K (2008) Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet 40:281–283. doi:10.1038/ng.89
Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, Benediktsdottir KR, Magnusdottir DN, Orlygsdottir G, Jakobsdottir M, Stacey SN, Sigurdsson A, Wahlfors T, Tammela T, Breyer JP, McReynolds KM, Bradley KM, Saez B, Godino J, Navarrete S, Fuertes F, Murillo L, Polo E, Aben KK, van Oort IM, Suarez BK, Helfand BT, Kan D, Zanon C, Frigge ML, Kristjansson K, Gulcher JR, Einarsson GV, Jonsson E, Catalona WJ, Mayordomo JI, Kiemeney LA, Smith JR, Schleutker J, Barkardottir RB, Kong A, Thorsteinsdottir U, Rafnar T, Stefansson K (2009) Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet 41:1122–1126. doi:10.1038/ng.448
Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ, Greenway SC, Stram DO, Le Marchand L, Kolonel LN, Frasco M, Wong D, Pooler LC, Ardlie K, Oakley-Girvan I, Whittemore AS, Cooney KA, John EM, Ingles SA, Altshuler D, Henderson BE, Reich D (2007) Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet 39:638–644. doi:10.1038/ng2015
Hammer MF, Behar DM, Karafet TM, Mendez FL, Hallmark B, Erez T, Zhivotovsky LA, Rosset S, Skorecki K (2009) Extended Y chromosome haplotypes resolve multiple and unique lineages of the Jewish priesthood. Hum Genet 126:707–717. doi:10.1007/s00439-009-0727-5
Jordan JJ, Hanlon AL, Al-Saleem TI, Greenberg RE, Tricoli JV (2001) Loss of the short arm of the Y chromosome in human prostate carcinoma. Cancer Genet Cytogenet 124:122–126. pii:S0165-4608(00)00340-X
Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF (2008) New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 18:830–838. doi:10.1101/gr.7172008
Kim W, Yoo TK, Kim SJ, Shin DJ, Tyler-Smith C, Jin HJ, Kwak KD, Kim ET, Bae YS (2007) Lack of association between Y-chromosomal haplogroups and prostate cancer in the Korean population. PLoS One 2:e172. doi:10.1371/journal.pone.0000172
Kote-Jarai Z, Olama AA, Giles GG, Severi G, Schleutker J, Weischer M, Campa D, Riboli E, Key T, Gronberg H, Hunter DJ, Kraft P, Thun MJ, Ingles S, Chanock S, Albanes D, Hayes RB, Neal DE, Hamdy FC, Donovan JL, Pharoah P, Schumacher F, Henderson BE, Stanford JL, Ostrander EA, Sorensen KD, Dork T, Andriole G, Dickinson JL, Cybulski C, Lubinski J, Spurdle A, Clements JA, Chambers S, Aitken J, Gardiner RA, Thibodeau SN, Schaid D, John EM, Maier C, Vogel W, Cooney KA, Park JY, Cannon-Albright L, Brenner H, Habuchi T, Zhang HW, Lu YJ, Kaneva R, Muir K, Benlloch S, Leongamornlert DA, Saunders EJ, Tymrakiewicz M, Mahmud N, Guy M, O’Brien LT, Wilkinson RA, Hall AL, Sawyer EJ, Dadaev T, Morrison J, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper CS, Lophatonanon A, Southey MC, Hopper JL, English DR, Wahlfors T, Tammela TL, Klarskov P, Nordestgaard BG, Roder MA, Tybjaerg-Hansen A, Bojesen SE, Travis R, Canzian F, Kaaks R, Wiklund F, Aly M, Lindstrom S, Diver WR, Gapstur S, Stern MC, Corral R, Virtamo J, Cox A, Haiman CA, Le Marchand L, Fitzgerald L, Kolb S et al (2011) Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet 43:785–791. doi:10.1038/ng.882
Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K (2000) Environmental and heritable factors in the causation of cancer–analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343:78–85. doi:10.1056/NEJM200007133430201
Lindstrom S, Adami HO, Adolfsson J, Wiklund F (2008) Y chromosome haplotypes and prostate cancer in Sweden. Clin Cancer Res 14:6712–6716. doi:10.1158/1078-0432.CCR-08-0658
Ma J, Li H, Giovannucci E, Mucci L, Qiu W, Nguyen PL, Gaziano JM, Pollak M, Stampfer MJ (2008) Prediagnostic body-mass index, plasma C-peptide concentration, and prostate cancer-specific mortality in men with prostate cancer: a long-term survival analysis. Lancet Oncol 9:1039–1047. doi:10.1016/S1470-2045(08)70235-3
Paracchini S, Pearce CL, Kolonel LN, Altshuler D, Henderson BE, Tyler-Smith C (2003) A Y chromosomal influence on prostate cancer risk: the multi-ethnic cohort study. J Med Genet 40:815–819
Rootsi S, Zhivotovsky LA, Baldovic M, Kayser M, Kutuev IA, Khusainova R, Bermisheva MA, Gubina M, Fedorova SA, Ilumae AM, Khusnutdinova EK, Voevoda MI, Osipova LP, Stoneking M, Lin AA, Ferak V, Parik J, Kivisild T, Underhill PA, Villems R (2007) A counter-clockwise northern route of the Y-chromosome haplogroup N from Southeast Asia towards Europe. Eur J Hum Genet 15:204–211. doi:10.1038/sj.ejhg.5201748
Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Page DC (2003) Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423:873–876. doi:10.1038/nature01723
Schumacher FR, Berndt SI, Siddiq A, Jacobs KB, Wang Z, Lindstrom S, Stevens VL, Chen C, Mondul AM, Travis RC, Stram DO, Eeles RA, Easton DF, Giles G, Hopper JL, Neal DE, Hamdy FC, Donovan JL, Muir K, Al Olama AA, Kote-Jarai Z, Guy M, Severi G, Gronberg H, Isaacs WB, Karlsson R, Wiklund F, Xu J, Allen NE, Andriole GL, Barricarte A, Boeing H, Bas Bueno-de-Mesquita H, Crawford ED, Diver WR, Gonzalez CA, Gaziano JM, Giovannucci EL, Johansson M, Le Marchand L, Ma J, Sieri S, Stattin P, Stampfer MJ, Tjonneland A, Vineis P, Virtamo J, Vogel U, Weinstein SJ, Yeager M, Thun MJ, Kolonel LN, Henderson BE, Albanes D, Hayes RB, Spencer Feigelson H, Riboli E, Hunter DJ, Chanock SJ, Haiman CA, Kraft P (2011) Genome-wide association study identifies new prostate cancer susceptibility loci. Hum Mol Genet 20:3867–3875. doi:10.1093/hmg/ddr295
Semino O, Magri C, Benuzzi G, Lin AA, Al-Zahery N, Battaglia V, Maccioni L, Triantaphyllidis C, Shen P, Oefner PJ, Zhivotovsky LA, King R, Torroni A, Cavalli-Sforza LL, Underhill PA, Santachiara-Benerecetti AS (2004) Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet 74:1023–1034. doi:10.1086/386295
Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, Kawaguchi T, Tsunoda T, Inazawa J, Kamatani N, Ogawa O, Fujioka T, Nakamura Y, Nakagawa H (2010) Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet 42:751–754. doi:10.1038/ng.635
The ATBC Cancer Prevention Study Group (1994) The alpha-tocopherol, beta-carotene lung cancer prevention study: design, methods, participant characteristics, and compliance. The ATBC Cancer Prevention Study Group. Ann Epidemiol 4:1–10
Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, Crenshaw A, Cancel-Tassin G, Staats BJ, Wang Z, Gonzalez-Bosquet J, Fang J, Deng X, Berndt SI, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cussenot O, Valeri A, Andriole GL, Crawford ED, Tucker M, Gerhard DS, Fraumeni JF Jr, Hoover R, Hayes RB, Hunter DJ, Chanock SJ (2008) Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet 40:310–315. doi:10.1038/ng.91
Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley RA, Oefner PJ, Cavalli-Sforza LL (2001) The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43–62. pii:S0003480001008582
Valeri A, Briollais L, Azzouzi R, Fournier G, Mangin P, Berthon P, Cussenot O, Demenais F (2003) Segregation analysis of prostate cancer in France: evidence for autosomal dominant inheritance and residual brother–brother dependence. Ann Hum Genet 67:125–137. pii:022
Vijayakumar S, Garcia D, Hensel CH, Banerjee M, Bracht T, Xiang R, Kagan J, Naylor SL (2005) The human Y chromosome suppresses the tumorigenicity of PC-3, a human prostate cancer cell line, in athymic nude mice. Genes Chromosomes Cancer 44:365–372. doi:10.1002/gcc.20250
Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N (2004) Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst 96:434–442
Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447:661–678. doi:10.1038/nature05911
Wiik K (2008) Where did European men come from? J Gen Genealogy 4:35–85
Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF Jr, Hoover R, Hunter DJ, Chanock SJ, Thomas G (2007) Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet 39:645–649. doi:10.1038/ng2022
Y Chromosome Consortium (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–48. doi:10.1101/gr.217602
Yeager M, Chatterjee N, Ciampa J, Jacobs KB, Gonzalez-Bosquet J, Hayes RB, Kraft P, Wacholder S, Orr N, Berndt S, Yu K, Hutchinson A, Wang Z, Amundadottir L, Feigelson HS, Thun MJ, Diver WR, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Crawford ED, Haiman CA, Henderson B, Kolonel L, Le Marchand L, Siddiq A, Riboli E, Key TJ, Kaaks R, Isaacs W, Isaacs S, Wiley KE, Gronberg H, Wiklund F, Stattin P, Xu J, Zheng SL, Sun J, Vatten LJ, Hveem K, Kumle M, Tucker M, Gerhard DS, Hoover RN, Fraumeni JF Jr, Hunter DJ, Thomas G, Chanock SJ (2009) Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet 41:1055–1057. doi:10.1038/ng.444
Acknowledgments
This study was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health (NIH). The authors acknowledge and thank both staff and participants in all studies for donating their time and making this study possible. The authors acknowledge support and constructive comments from Dr. Stephen J. Chanock, LTG, DCEG, National Cancer Institute. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Conflict of interest
All authors report no financial interests or potential conflicts of interests.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Z. Wang and H. Parikh are co-first authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Wang, Z., Parikh, H., Jia, J. et al. Y chromosome haplogroups and prostate cancer in populations of European and Ashkenazi Jewish ancestry. Hum Genet 131, 1173–1185 (2012). https://doi.org/10.1007/s00439-012-1139-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-012-1139-5