Abstract
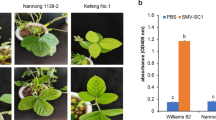
Soybean mosaic virus (SMV) is one of the most devastating pathogens for soybeans in China. Among the country-wide 22 strains, SC5 dominates in Huang-Huai and Changjiang valleys. For controlling its damage, the resistance gene was searched through Mendelian inheritance study, gene fine-mapping, and candidate gene analysis combined with qRT-PCR (quantitative real-time polymerase chain reaction) analysis. The parents F1, F2, and RILs (recombinant inbred lines) of the cross Kefeng-1 (Resistance, R) × NN1138-2 (Susceptible, S) were used to examine the inheritance of SC5-resistance. The F1 was resistant and the F2 and RILs segregated in a 3R:1S and 1R:1S ratio, respectively, indicating a single dominant gene conferring the Kefeng-1 resistance. Subsequently, the genomic region conferring the resistance was found in “Bin 352–Bin353 with 500 kb” on Chromosome 2 using the phenotyping data of the 427 RILs and a high-density genetic map with 4703 bin markers. In the 500 kb genomic region, 38 putative genes are contained. The association analysis between the SNPs in a putative gene and the resistance phenotype for the 427 RILs prioritized 11 candidate genes using Chi-square criterion. The expression levels of these genes were tested by qRT-PCR. On infection with SC5, 7 out of the 11 genes had differential expression in Kefeng-1 and NN1138-2. Furthermore, integrating SNP-phenotype association analysis with qRT-PCR expression profiling analysis, Glyma02g13495 was found the most possible candidate gene for SC5-resistance. This finding can facilitate the breeding for SC5-resistance through marker-assisted selection and provide a platform to gain a better understanding of SMV-resistance gene system in soybean.


Similar content being viewed by others
References
Cavanagh CR, Chao S, Wang S, Huang BE, Stephen S, Kiani S, Forrest K, Saintenac C, Brown-Guedira GL, Akhunova A, See D, Bai GH, Pumphrey M, Tomar L, Wong D, Kong S, Reynolds M, Sliva ML, Bockelman H, Talbert L, Anderson JA, Dreisigacker S, Baenzigr S, Carter A, Korzun V, Morrell PL, Dubcovsky J, Morell MK, Sorrells ME, Hayden MJ, Akhunov E (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci 110(20):8057–8062
Chanclud E, Kisiala A, Emery NR, Chalvon V, Ducasse A, Romiti-Michel C, Gravot A, Kroj T, Morel JB (2016) Cytokinin production by the rice blast fungus is a pivotal requirement for full virulence. PLos Pathog 12(2):e1005457. doi:10.1371/journal.ppat.1005457
Chen P, Choi CW (2007) Characterization of genetic interaction between soybean and soybean mosaic virus. In: Rao GP (ed) Molecular diagnosis of plant viruses. Studium Press, Houston, pp 389–422
Chen YX, Xue BD, Hu YZ, Fang ZD (1986) Identification of two new strains of soybean mosaic virus. Acta Phytophylacica Sin 13:221–226
Chen Z, Wang B, Dong X, Liu H, Ren L, Chen J, Hauck A, Song W, Lai JS (2014) An ultra-high density bin-map for rapid QTL mapping for tassel and ear architecture in a large F2 maize population. BMC Genom 15:433
Cho EK, Goodman RM (1979) Strains of soybean mosaic virus: classification based on virulence in resistant soybean cultivars. Phytopathology 69:467–470
Cho EK, Goodman RM (1982) Evaluation of resistance in soybeans to soybean mosaic virus strains. Crop Sci 22:1133–1136
Choi J, Huh SU, Kojima M, Sakakibara H, Paek KH, Hwang I (2010) The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev Cell 19:284–295
Choi J, Choi D, Lee S, Ryu CM, Hwang I (2011) Cytokinins and plant immunity: old foes or new friends. Trends Plant Sci 16:388–394
Chung B, Lee S, Cho E, Park H (1974) Identification of soybean viruses and soybean varietal reactions. Annu Rep Circ 26:137–145
Doyle JJ (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Fu SX, Zhan Y, Zhi HJ, Gai JY, Yu DY (2006) Mapping of SMV resistance gene Rsc-7 by SSR markers in soybean. Genetica 128:63–69
Ganal MW, Altmann T, Roder MS (2009) SNP identification in crop plants. Curr Opin Plant Biol 12:211–217
Guo DQ, Zhi HJ, Wang YW, Gai JY, Zhou XA, Yang CL, Li K, Li HC (2005) Identification and distribution of soybean mosaic virus strains in middle and northern Huang Huai region of China. Soybean Sci 27:64–68
Guo DQ, Wang YW, Zhi HJ, Gai JY, Li HC, Li K (2007) Inheritance and gene mapping of resistance to SMV strain group SC-13 in soybean. Soybean Sci 26:21–24
Hayes AJ, Ma G, Buss GR, Maroof M (2000) Molecular marker mapping of Rsv4, a gene conferring resistance to all known strains of soybean mosaic virus. Crop Sci 40:1434–1437
Hill J (1999) Soybean mosaic. In: Hartman GL, Sinclair JB, Rupe JC (eds) Compendium of soybean diseases, 4th edn. American Phytopathological Society, St. Paul, pp 70–71
Huang X, Feng Q, Qian Q, Zhao Q, Wang L, Wang A, Guan J, Fan D, Weng Q, Huang T (2009) High-throughput genotyping by whole-genome resequencing. Genome Res 19:1068–1076
Hwang TY, Moon JK, Yu S, Yang K, Mohankumar S, Yu YH, Lee YH, Kim HS, Kim HM, Maroof MAS (2006) Application of comparative genomics in developing molecular markers tightly linked to the virus resistance gene Rsv4 in soybean. Genome 49:380–388
Hyten DL, Choi IY, Song Q, Specht JE, Carter TE, Shoemaker RC, Hwang EY, Matukumalli LK, Cregan PB (2010) A high density integrated genetic linkage map of soybean and the development of a 1536 universal soy linkage panel for quantitative trait locus mapping. Crop Sci 50:960–968
Ilut DC, Lipka AE, Jeong N, Bae DN, Kim DH, Kim JH, Redekar N, Yang K, Park W, Kang ST, Kim N, Moon JK, Saghai Maroof MA, Gore MA, Jeong SC (2015) Identification of haplotypes at the Rsv4 genomic region in soybean associated with durable resistance to soybean mosaic virus. Theor Appl Genet 9:1–16
Jeong S, Maroof S (2004) Detection and genotyping of SNPs tightly linked to two disease resistance loci, Rsv1 and Rsv3, of soybean. Plant Breed 123:305–310
Jones E, Chu WC, Ayele M, Ho J, Bruggeman E, Yourstone K, Rafalski A, Smith OS, McMullen MD, Bezawada C (2009) Development of single nucleotide polymorphism (SNP) markers for use in commercial maize (Zea mays L.) germplasm. Mol Breed 24:165–176
Kale SM, Jaganathan D, Ruperao P, Chen C, Punna R, Kudapa H, Thudi M, Roorkiwal M, Katta MA, Doddamani D (2015) Prioritization of candidate genes in “QTL-hotspot” region for drought tolerance in chickpea (Cicer arietinum L.). Sci Rep. doi:10.1038/srep15296
Kiba T, Naitou T, Koizumi N, Yamashino T, Sakakibara H, Mizuno T (2005) Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His → Asp phosphorelay circuitry. Plant Cell Physiol 46:339–355
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultra fast tool for short read alignment. Bioinformatics 25:1966–1967
Li K, Yang QH, Zhi HJ, Gai JY (2010) Identification and distribution of soybean mosaic virus strains in southern China. Plant Dis 94:351–357
Li N, Yin JL, Li C, Wang DG, Yang YQ, Karthikeyan A, Luan HX, Zhi HJ (2016) NB-LRR gene family required for Rsc4-mediated resistance to soybean mosaic virus. Crop Pasture Sci 67: 541–552
Liu JZ, Fang Y, Pang H (2016) The current status of the soybean—soybean mosaic virus (SMV) pathosystem. Front Microbiol 7:1906. doi:10.3389/fmicb.2016.01906
Ma Y, Wang DG, Li HC, Zheng GJ, Yang YQ, Li HC, Zhi HJ (2011) Fine mapping of the RSC14Q locus for resistance to soybean mosaic virus in soybean. Euphytica 181:127–135
Maroof M, Tucker DM, Skoneczka JA, Bowman BC, Tripathy S, Tolin SA (2010) Fine mapping and candidate gene discovery of the soybean mosaic virus resistance gene, Rsv4. Plant Genome 3:14–22
McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, Zeller G, Clark RM, Hoen DR, Bureau TE (2009) Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc Natl Acad Sci 106:12273–12278
Meng S, He J, Zhao T, Xing G, Li Y, Yang S, Lu J, Wang Y, Gai J (2016) Detecting the QTL allele system of seed isoflavone content in Chinese soybean landrace population for optimal cross design and gene system exploration. Theor Appl Genet 129:1557–1576
Michelmore RW, Christopoulou M, Caldwell KS (2013) Impacts of resistance gene genetics, function, and evolution on a durable future. Annu Rev Phytopathol 51:291–319
Obrien JA, Benkova E (2013) Cytokinin cross-talking during biotic and abiotic stress responses. Front Plant Sci. doi:10.3389/fpls.2013.00451
Poland JA, Rife TW (2012) Genotyping-by-sequencing for plant breeding and genetics. Plant Genome 5:92–102
Poland JA, Brown PJ, Sorrells ME, Jannink JL (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7:e32253
Pu Z, Cao Q, Fang D, Xue B, Fang Z (1982) Identification of strains of soybean mosaic virus. Acta Phytophylacica Sin 9:15–19
Qi X, Li M, Xie M, Liu X, Ni M, Shao G, Song C, Yim AK, Tao Y, Wong FL (2014) Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat Commun 5:4340
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Song Q, Hyten DL, Jia G, Quigley CV, Fickus EW, Nelson RL, Cregan PB (2013) Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS One 8:e54985
Suh SJ, Bowman BC, Jeong N, Yang K, Kastl C, Tolin SA, Maroof M, Jeong SC (2011) The Rsv3 locus conferring resistance to soybean mosaic virus is associated with a cluster of coiled-coil nucleotide-binding leucine-rich repeat genes. Plant Genome 4:55–64
Takahashi K, Tanaka T, Iida W, Tsuda Y (1980) Studies on virus diseases and causal viruses of soybean in Japan. Bull Tohoku Natl Agric Exp Stn 62:1–130
Thudi M, Upadhyaya HD, Rathore A, Gaur PM, Krishnamurthy L, Roorkiwal M, Nayak SN, Chaturvedi SK, Basu PS, Gangarao NV, Fikre A, Kimurto P, Sharma PC, Sheshashayee MS, Tobita S, Kashiwagi J, Ito O, Killian A, Varshney RK (2014) Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. PLoS One 9:e96758
Tian H, Wang F, Zhao J, Yi H, Wang L, Wang R, Yang Y, Song W (2015) Development of maize SNP3072, a high-throughput compatible SNP array, for DNA fingerprinting identification of Chinese maize varieties. Mol Breed 35:1–11
USDA (2015) World Agricultural Supply and Demand Estimates
Van Ooijen JW (2006) JoinMap 4.0, software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V., Wageningen
Varshney RK, Nayak SN, May GD, Jackson SA (2009) Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol 27:522–530
Wang X, Gai J, Pu Z (2003) Classification and distribution of strains of soybean mosaic virus middle and lower Huang-Huai and Changjiang valleys. Soybean Sci 22:102–107
Wang Y, Dong F, Wang X, Yang Y, Yu D, Gai J, Wu X, He C, Zhang J, Chen S (2004) Mapping of five genes resistant to SMV strains in soybean. Acta Genet Sin 31:87–90
Wang Y, Zhi H, Guo D, Gai J, Chen Q, Li K, Li H (2005) Classification and distribution of strains of Soybean mosaic virus in northern China spring planting soybean region. Soybean Sci 24:263–268
Wang DG, Ma Y, Liu N, Yang ZL, Zheng GJ, Zhi HJ (2011a) Fine mapping of resistance to soybean mosaic virus strain SC4 in soybean based on genomic-SSR markers. Plant Breed 130:653–659
Wang DG, Ma Y, Yang YQ, Liu N, Li CY, Song YP, Zhi HJ (2011b) Fine mapping and analyses of RSC8 resistance candidate genes to soybean mosaic virus in soybean. Theor Appl Genet 122:555–565
Wang D, Tian Z, Li K, Li H, Huang Z, Hu G, Zhang L, Zhi H (2013) Identification and variation analysis of soybean mosaic virus strains in Shandong, Henan and Anhui provinces of China. Soybean Sci 32:806–809
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L (2014a) Characterization of polyploid wheat genomic diversity using a high-density 90000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796
Wang D, Li H, Zhi H, Tian Z, Hu C, Hu G, Huang Z, Zhang L (2014b) Identification of strains and screening of resistance resources to soybean mosaic virus in Anhui province. Soybean Sci 36(3):374–379
Wilkinson PA, Winfield MO, Barker GL, Allen AM, Burridge A, Coghill JA (2012) Cereals DB 2.0: an integrated resource for plant breeders and scientists. BMC Bioinform 13:219
Xie W, Feng Q, Yu H, Huang X, Zhao Q, Xing Y, Yu S, Han B, Zhang Q (2010) Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proc Natl Acad Sci 107:10578–10583
Xu X, Zeng L, Tao Y, Vuong T, Wan J, Boerma R, Noe J, Li Z, Finnerty S, Pathan SM (2013) Pinpointing genes underlying the quantitative trait loci for root-knot nematode resistance in palaeopolyploid soybean by whole genome resequencing. Proc Natl Acad Sci 110:13469–13474
Xu J, Yuan Y, Xu Y, Zhang G, Guo X, Wu F, Wang Q, Rong T, Pan G, Cao M (2014) Identification of candidate genes for drought tolerance by whole-genome resequencing in maize. BMC Plant Biol 14:1–15
Yan H, Wang H, Cheng H, Hu Z, Chu S, Zhang G, Yu D (2015) Detection and fine-mapping of SC7 resistance genes via linkage and association analysis in soybean. J Integr Plant Biol 57:722–729
Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo Z, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS (2010) Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329:75–78
Yonemaru J, Ebana K, Yano M (2014) HapRice, an SNP haplotype database and a web tool for rice. Plant Cell Physiol 55:e9
Zhan Y, Zhi H, Yu D, Gai J (2006) Identification and distribution of SMV strains in Huang-Huai valleys. Sci Agric Sin 39:2009–2015
Zheng GJ, Yang YQ, Ma Y, Yang XF, Chen SY, Ren R, Wang DG, Yang ZL, Zhi HJ (2014) Fine mapping and candidate gene analysis of resistance gene R SC3Q to soybean mosaic virus in Qihuang No. 1. J Integr Agric 13:2608–2615
Acknowledgements
This work was financially supported through grants from the China National Key Basic Research Program (2011CB1093), the China National High-tech R&D Program (2012AA101106), the Natural Science Foundation of China (31671718), the MOE111 Project (B08025), the MOE Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT13073), the MOA CARS-04 program, the Jiangsu Higher Education PAPD Program, the Fundamental Research Funds for the Central Universities, and the Jiangsu JCIC-MCP. The funders had no role in work design, data collection and analysis, or decision and preparation of the manuscript.
Author contributions
JG, SC, and HZ designed the methods and experiments. AK, JH, RR, and LC conducted SMV inoculations and resistance evaluation, and managed field work. AK and KL performed the qRT-PCR analysis, A.K analyzed the data. AK, KL, and JG drafted the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors, AK, JH, RR, LC, SC, HZ, SC, and JG declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
Additional information
Communicated by S. Hohmann.
A. Karthikeyan and K. Li contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Karthikeyan, A., Li, K., Jiang, H. et al. Inheritance, fine-mapping, and candidate gene analyses of resistance to soybean mosaic virus strain SC5 in soybean. Mol Genet Genomics 292, 811–822 (2017). https://doi.org/10.1007/s00438-017-1310-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-017-1310-8