Abstract
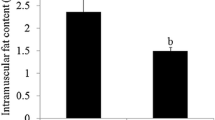
Skeletal muscle fibers are mainly categorized into red and white fiber types, and the ratio of red/white fibers within muscle mass plays a crucial role in meat quality such as tenderness and flavor. To better understand the molecular difference between the two muscle fibers, this study takes advantage of RNA-seq to compare differences in the transcriptome between extensor digitorum longus (EDL; white fiber) and soleus (Sol; red fiber) muscles of large white pigs. In total, 89,658,562 and 46,723,568 raw reads from EDL and Sol were generated, respectively. Comparison between the two transcriptomes revealed 561 differentially expressed genes, with 408 displaying higher and 153 lower levels of expression in Sol. Quantitative real-time polymerase chain reaction validated the differential expression of nine genes. Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis discovered several differentially enriched biological functions and processes of the two muscles. Moreover, transcriptome comparison between EDL and Sol identified many muscle-related genes (CSRP3, ACTN2, MYL1, and MYH6) and pathways related to myofiber formation, such as focal adhesion, tight junction formation, extracellular matrix (ECM)–receptor pathway, calcium signaling, and Wnt signaling. In addition, 58,362 and 58,359 single nucleotide polymorphisms were identified in EDL and Sol, respectively, and the sequence of 9069 genes was refined at the 5′, 3′ or both ends. Numerous novel transcripts and alternatively spliced RNAs were also identified. Our transcriptome analysis constitutes valuable sequence resource for uncovering important genes and pathways involved in muscle fiber type determination, and might help further our understanding of the molecular mechanisms in different types of muscle.






Similar content being viewed by others
Abbreviations
- EDL:
-
Extensor digitorum longus
- Sol:
-
Soleus
- DEGs:
-
Differentially expressed genes
- GO:
-
Gene ontology
- SNPs:
-
Single nucleotide polymorphisms
- CPC:
-
Coding potential calculator
- FDR:
-
False discovery rate
- AS:
-
Alternative splicing
References
Anakwe K, Robson L, Hadley J, Buxton P, Church V, Allen S, Hartmann C, Harfe B, Nohno T, Brown AMC, Evans DJR, Francis-West P (2003) Wnt signalling regulates myogenic differentiation in the developing avian wing. Development 130:3503–3514
Arber S, Halder G, Caroni P (1994) Muscle lim protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell 79:221–231
Bai Q, McGillivray C, da Costa N, Dornan S, Evans G, Stear MJ, Chang KC (2003) Development of a porcine skeletal muscle cDNA microarray: analysis of differential transcript expression in phenotypically distinct muscles. BMC Genom 4:8
Chin ER, Olson EN, Richardson JA, Yano Q, Humphries C, Shelton JM, Wu H, Zhu WG, Bassel-Duby R, Williams RS (1998) A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Gene Dev 12:2499–2509
Choi YM, Lee SH, Choe JH, Rhee MS, Lee SK, Joo ST, Kim BC (2010) Protein solubility is related to myosin isoforms, muscle fiber types, meat quality traits, and postmortem protein changes in porcine longissimus dorsi muscle. Livest Sci 127:183–191
Choi YM, Nam KW, Choe JH, Ryu YC, Wick MP, Lee K, Kim BC (2013) Growth, carcass, fiber type, and meat quality characteristics in large white pigs with different live weights. Livest Sci 155:123–129
Garcia de la Serrana D, Estevez A, Andree K, Johnston IA (2012) Fast skeletal muscle transcriptome of the gilthead sea bream (Sparus aurata) determined by next generation sequencing. BMC Genom 13:181
Geiger B, Bershadsky A (2002) Exploring the neighborhood: adhesion-coupled cell mechanosensors. Cell 110:139–142
Huang S, Zhang J, Li R, Zhang W, He Z, Lam TW, Peng Z, Yiu SM (2011) SOAPsplice: genome-wide ab initio detection of splice junctions from RNA-seq data. Front Genet 2:46
Lee EA, Kim JM, Lim KS, Ryu YC, Jeon WM, Hong KC (2012) Effects of variation in porcine MYOD1 gene on muscle fiber characteristics, lean meat production, and meat quality traits. Meat Sci 92:36–43
Lefaucheur L, Milan D, Ecolan P, Le Callennec C (2004) Myosin heavy chain composition of different skeletal muscles in large white and Meishan pigs. J Anim Sci 82:1931–1941
Leitinger B (2011) Transmembrane collagen receptors. Annu Rev Cell Dev Biol 27:265–290
Lepetit J (2007) A theoretical approach of the relationships between collagen content, collagen cross-links and meat tenderness. Meat Sci 76:147–159
Li RQ, Li YR, Fang XD, Yang HM, Wang J, Kristiansen K, Wang J (2009a) SNP detection for massively parallel whole-genome resequencing. Genome Res 19:1124–1132
Li RQ, Yu C, Li YR, Lam TW, Yiu SM, Kristiansen K, Wang J (2009b) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967
Li Y, Xu ZY, Li HY, Xiong YZ, Zuo B (2010) Differential transcriptional analysis between red and white skeletal muscle of Chinese Meishan pigs. Int J Biol Sci 6:350–360
Lim KS, Lee SH, Lee EA, Kim JM, Hong KC (2015) Effects of intergenic single nucleotide polymorphisms in the fast myosin heavy chain cluster on muscle fiber characteristics and meat quality in Berkshire pigs. Meat Sci 110:224–229
MacArthur DG, Seto JT, Raftery JM, Quinlan KG, Huttley GA, Hook JW, Lemckert FA, Kee AJ, Edwards MR, Berman Y, Hardeman EC, Gunning PW, Easteal S, Yang N, North KN (2007) Loss of ACTN3 gene function alters mouse muscle metabolism and shows evidence of positive selection in humans. Nat Genet 39:1261–1265
Maltin C, Balcerzak D, Tilley R, Delday M (2003) Determinants of meat quality: tenderness. P Nutr Soc 62:337–347
Mills MA, Yang N, Weinberger RP, Vander Woude DL, Beggs AH, Easteal S, North KN (2001) Differential expression of the actin-binding proteins, alpha-actinin-2 and-3, in different species: implications for the evolution of functional redundancy. Hum Mol Genet 10:1335–1346
Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Naya FJ, Mercer B, Shelton J, Richardson JA, Williams RS, Olson EN (2000) Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J Biol Chem 275:4545–4548
Palstra AP, Beltran S, Burgerhout E, Brittijn SA, Magnoni LJ, Henkel CV, Jansen HJ, van den Thillart GE, Spaink HP, Planas JV (2013) Deep RNA sequencing of the skeletal muscle transcriptome in swimming fish. PLoS One 8:e53171
Rehfeldt C, Henning M, Fiedler I (2008) Consequences of pig domestication for skeletal muscle growth and cellularity. Livest Sci 116:30–41
Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, Kam Z, Geiger B, Bershadsky AD (2001) Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol 153:1175–1185
Roberts A, Pimentel H, Trapnell C, Pachter L (2011) Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27:2325–2329
Ruusunen M, Puolanne E (2004) Histochemical properties of fibre types in muscles of wild and domestic pigs and the effect of growth rate on muscle fibre properties. Meat Sci 67:533–539
Ruusunen M, Puolanne E, Sevon-Aimonen ML, Partanen K, Voutila L, Niemi J (2012) Carcass and meat quality traits of four different pig crosses. Meat Sci 90:543–547
Schneider AG, Sultan KR, Pette D (1999) Muscle LIM protein: expressed in slow muscle and induced in fast muscle by enhanced contractile activity. Am J Physiol 276:C900–C906
Sebastian S, Faralli H, Yao ZZ, Rakopoulos P, Palii C, Cao Y, Singh K, Liu QC, Chu A, Aziz A, Brand M, Tapscott SJ, Dilworth FJ (2013) Tissue-specific splicing of a ubiquitously expressed transcription factor is essential for muscle differentiation. Gene Dev 27:1247–1259
Sethi JK, Vidal-Puig A (2010) Wnt signalling and the control of cellular metabolism. Biochem J 427:1–17
Takata H, Terada K, Oka H, Sunada Y, Moriguchi T, Nohno T (2007) Involvement of Wnt4 signaling during myogenic proliferation and differentiation of skeletal muscle. Dev Dyn 236:2800–2807
Tiso N, Majetti N, Stanchi F, Rampazzo A, Zimbello R, Nava A, Danieli GA (1999) Fine mapping and genomic structure of ACTN2, the human gene coding for the sarcomeric isoform of alpha-actinin-2, expressed in skeletal and cardiac muscle. Biochem Bioph Res Commun 265:256–259
von Maltzahn J, Chang NC, Bentzinger CF, Rudnicki MA (2012) Wnt signaling in myogenesis. Trends Cell Biol 22:602–609
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63
Wang LK, Feng ZX, Wang X, Wang XW, Zhang XG (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138
Wei B, Jin JP (2011) Troponin T isoforms and posttranscriptional modifications: evolution, regulation and function. Arch Biochem Biophys 505:144–154
Weiskirchen R, Gunther K (2003) The CRP/MLP/TLP family of LIM domain proteins: acting by connecting. BioEssays 25:152–162
Wu H, Naya FJ, McKinsey TA, Mercer B, Shelton JM, Chin ER, Simard AR, Michel RN, Bassel-Duby R, Olson EN, Williams RS (2000) MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J 19:1963–1973
Xu XW, Qiu HF, Du ZQ, Fan B, Rothschild MF, Yuan F, Liu B (2010) Porcine CSRP3: polymorphism and association analyses with meat quality traits and comparative analyses with CSRP1 and CSRP2. Mol Biol Rep 37:451–459
Acknowledgments
We thank the Beijing Genomics Institute (BGI)-Shenzhen for helping us with technical assistance in DGE and Edanz for providing language editing service.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None of the authors declare any conflict of interest.
Ethical approval
The large white pigs were supplied by the Northwest A&F University Experiment Station (Yangling, China). Experiments were handled in accordance with the guidelines of the Administration of Affairs Concerning Experimental Animals and authorized by the Institutional Animal Care and Use Committee of the College of Animal Science and Technology, Northwest A&F University, Shaanxi, China. All applicable international, national, and institutional guidelines for the care and use of animals were followed.
Funding
This study was supported by the National Basic Research Program of China (Grant No. 2015CB943102), the National Natural Science Foundation of China (Grant No. 31272410), and the Scientific and Technological Innovation Project for Youths of Northwest A&F University (Grant No. ZD2012007). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional information
Communicated by S. Hohmann.
Jiayu Zhu and Xin’e Shi contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhu, J., Shi, X., Lu, H. et al. RNA-seq transcriptome analysis of extensor digitorum longus and soleus muscles in large white pigs. Mol Genet Genomics 291, 687–701 (2016). https://doi.org/10.1007/s00438-015-1138-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-015-1138-z