Abstract
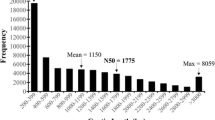
Welsh onion (Allium fistulosum L.) has long been cultivated as a vegetable and spice for its flavor and aroma. However, transcriptomic and genomic data for A. fistulosum remain scarce. The goal of this study was to generate transcript sequences for functional genomic analyses, and identify genes potentially involved in sulfur, selenium, and vitamin metabolism. In total, 53,378,674 high-quality reads were generated, and de novo assembly resulted in 103,286 contigs and 53,837 unigenes. The average unigene length was 619 bp with an N50 of 832 bp. Similarity searches revealed that 36,155 sequences were similar to those of known proteins in public databases. Of these, 35,250 unigenes sequences were significantly similar to sequences in the NCBI non-redundant protein database and 22,804 were annotated in the Swiss-Prot database. Additionally, 13,125 and 26,660 unigenes were annotated in the Cluster of Orthologous Group and Gene Ontology databases, respectively. A total of 20,680 unigenes were classified into 128 pathways via functional annotation against the Kyoto Encyclopedia of Genes and Genomes pathway database. Key enzymes involved in sulfur and selenium metabolism were also identified. Additionally, our transcriptome revealed a number of unigenes encoding important enzymes involved in vitamin metabolism. We also identified 2014 simple sequence repeats in 1892 unigenes. This transcriptome analysis provides valuable information to further our understanding of the molecular mechanisms regulating the biosynthesis of organic sulfur compounds. The detected simple sequence repeats may facilitate marker-assisted selection in Welsh onion breeding experiments.





Similar content being viewed by others
References
Ashrafi H, Hill T, Stoffel K, Kozik A, Yao J, Chin-Wo SR, Van Deynze A (2012) De novo assembly of the pepper transcriptome (Capsicum annuum): a benchmark for in silico discovery of SNPs. SSRs and candidate genes. BMC Genomics 13:571
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Coolong TW, Randle WM (2003) Temperature influences flavor intensity and quality inGranex 33′onion. J Am Soc Hortic Sci 128:176–181
Diplock AT (1994) Antioxidants and disease prevention. Mol Aspects of Med 15:293–376
Dong Y, Cheng Z, Meng H, Liu H, Wu C, Khan AR (2013) The effect of cultivar, sowing date and transplant location in field on bolting of Welsh onion (Allium fistulosum L.). BMC Plant Biol 13:154
Dong S, Liu Y, Niu J, Ning Y, Lin S, Zhang Z (2014) De novo transcriptome analysis of the Siberian apricot (Prunus sibirica L.) and search for potential SSR markers by 454 pyrosequencing. Gene 544:220–227
Du F, Wu Y, Zhang L, Li XW, Zhao XY, Wang WH, Xia YP (2014) De novo assembled transcriptome analysis and SSR marker development of a mixture of six tissues from Lilium Oriental hybrid ‘Sorbonne’. Plant Mol Biol Rep 2014:1–13
Garg R, Patel RK, Tyagi AK, Jain M (2011) De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res 18:53–63
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Hughes J, Tregova A, Tomsett AB, Jones MG, Cosstick R, Collin HA (2005) Synthesis of the flavour precursor, alliin, in garlic tissue cultures. Phytochemistry 66:187–194
Jones MG, Hughes J, Tregova A, Milne J, Tomsett AB, Collin HA (2004) Biosynthesis of the flavour precursors of onion and garlic. J Exp Bot 55:1903–1918
Kamenetsky R, Faigenboim A, Shemesh-Mayer E, Ben-Michael T, Gershberg C, Kimhi S, Sherman A (2015) Integrated transcriptome catalogue and organ-specific profiling of gene expression in fertile garlic (Allium sativum L.). BMC Genomics 16:12
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484
Kim S, Kim MS, Kim YM, Yeom SI, Cheong K, Kim KT, Choi D (2014) Integrative structural annotation of de novo RNA-Seq provides an accurate reference gene set of the enormous genome of the onion (Allium cepa L.). DNA Res 22:19–27
Kopriva S (2006) Regulation of sulfate assimilation in Arabidopsis and beyond. Ann Bot Lond 97:479–495
Kuhl JC, Cheung F, Yuan Q, Martin W, Zewdie Y, McCallum J, Havey MJ (2004) A unique set of 11,008 onion expressed sequence tags reveals expressed sequence and genomic differences between the monocot orders Asparagales and Poales. Plant Cell 16:114–125
Kumar S, Shah N, Garg V, Bhatia S (2014) Large scale in silico identification and characterization of simple sequence repeats (SSRs) from de novo assembled transcriptome of Catharanthus roseus (L.) G. Don. Plant Cell Rep 33:905–918
Labani RM, Elkington TT (1987) Nuclear DNA variation in the genus Allium L. (Liliaceae). Heredity 59:119–128
Li D, Deng Z, Qin B, Liu X, Men Z (2012) De novo assembly and characterization of bark transcriptome using Illumina sequencing and development of EST-SSR markers in rubber tree (Hevea brasiliensis Muell. Arg.). BMC Genomics 13:192
Liu T, Zhu S, Tang Q, Chen P, Yu Y, Tang S (2013) De novo assembly and characterization of transcriptome using Illumina paired-end sequencing and identification of CesA gene in ramie (Boehmeria nivea L. Gaud). BMC Genomics 14:125
McCallum J, Clarke A, Pither-Joyce M, Shaw M, Butler R, Brash D, Havey MJ (2006) Genetic mapping of a major gene affecting onion bulb fructan content. Theor Appl Genet 112:958–967
McCallum J, Pither-Joyce M, Shaw M, Kenel F, Davis S, Butler R, Havey MJ (2007) Genetic mapping of sulfur assimilation genes reveals a QTL for onion bulb pungency. Theor Appl Genet 114:815–822
McCallum J, Thomas L, Shaw M, Pither-Joyce M, Leung S, Cumming M, McManus MT (2011) Genotypic variation in the sulfur assimilation and metabolism of onion (Allium cepa L.) I. Plant composition and transcript accumulation. Phytochemistry 72:882–887
McManus MT, Joshi S, Searle B, Pither-Joyce M, Shaw M, Leung S, McCallum J (2012) Genotypic variation in sulfur assimilation and metabolism of onion (Allium cepa L.) III. Characterization of sulfite reductase. Phytochemistry 83:34–42
Mudalkar S, Golla R, Ghatty S, Reddy AR (2014) De novo transcriptome analysis of an imminent biofuel crop, Camelina sativa L. using Illumina GAIIX sequencing platform and identification of SSR markers. Plant Mol Biol 84:159–171
Novoselov SV, Rao M, Onoshko NV, Zhi H, Kryukov GV, Xiang Y, Gladyshev VN (2002) Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii. EMBO J 21:3681–3693
Ohri D, Fritsch RM, Hanelt P (1998) Evolution of genome size in Allium (Alliaceae). Plant Syst Evol 210:57–86
Pilon-Smits EA, Hwang S, Lytle CM, Zhu Y, Tai JC, Bravo RC, Terry N (1999) Overexpression of ATP sulfurylase in Indian mustard leads to increased selenate uptake, reduction, and tolerance. Plant Physiol 119:123–132
Randle WM (2000) Increasing nitrogen concentration in hydroponic solutions affects onion flavor and bulb quality. J Am Soc Hortic Sci 125(2):254–259
Ricroch A, Yockteng R, Brown SC, Nadot S (2005) Evolution of genome size across some cultivated Allium species. Genome 48:511–520
Salgado LR, Koop DM, Pinheiro DG, Rivallan R, Le Guen V, Nicolás MF, Garcia D (2014) De novo transcriptome analysis of Hevea brasiliensis tissues by RNA-seq and screening for molecular markers. BMC Genomics 15:236
Shahin A, van Kaauwen M, Esselink D, Bargsten JW, van Tuyl JM, Visser RG, Arens P (2012) Generation and analysis of expressed sequence tags in the extreme large genomes Lilium and Tulipa. BMC Genomics 13:640
Sors TG, Ellis DR, Na GN, Lahner B, Lee S, Leustek T, Salt DE (2005) Analysis of sulfur and selenium assimilation in Astragalus plants with varying capacities to accumulate selenium. Plant J 42:785–797
Stadtman TC (1996) Selenocysteine. Annu Rev Biochem 65:83–100
Sun X, Zhou S, Meng F, Liu S (2012) De novo assembly and characterization of the garlic (Allium sativum) bud transcriptome by Illumina sequencing. Plant Cell Rep 31:1823–1828
Tsukazaki H, Yaguchi S, Shusei Sato S, Hirakawa H, Katayose Y, Kanamori H, Wako T (2015) Development of transcriptome shotgun assembly-derived markers in bunching onion (Allium fistulosum). Mol Breed 35:55
Wang G, Du X, Ji J, Guan C, Li Z, Josine TL (2015) De novo characterization of the Lycium chinense Mill. leaf transcriptome and analysis of candidate genes involved in carotenoid biosynthesis. Gene 555:458–463
Xie F, Burklew CE, Yang Y, Liu M, Xiao P, Zhang B, Qiu D (2012) De novo sequencing and a comprehensive analysis of purple sweet potato (Impomoea batatas L.) transcriptome. Planta 236:101–113
Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:W293–W297
Yuan S, Ge L, Liu C, Ming J (2013) The development of EST-SSR markers in Lilium regale and their cross-amplification in related species. Euphytica 189:393–419
Zhang MF, Jiang LM, Zhang DM, Jia GX (2015) De novo transcriptome characterization of Lilium ‘Sorbonne’ and key enzymes related to the flavonoid biosynthesis. Mol Genet Genomics 290:399–412
Zhong M, Liu B, Wang X, Liu L, Lun Y, Li X, Huang M (2013) De novo characterization of Lentinula edodes C91–3 transcriptome by deep Solexa sequencing. Biochem Biophys Res Commun 431:111–115
Zhou SM, Chen LM, Liu SQ, Wang XF, Sun XD (2015) De novo assembly and annotation of the Chinese chive (Allium tuberosum Rottler ex Spr.) transcriptome using the Illumina platform. PLoS One 10:7
Zhu L, Zhang Y, Guo W, Xu XJ, Wang Q (2014) De novo assembly and characterization of Sophora japonica transcriptome using RNA-seq. BioMed Res Int 2014:2014
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
We declare that we have no financial or personal relationships with other people or organizations that can inappropriately influence our work. There is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in our manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (31372084) and by the Chinese National Department Public Benefit Research Foundation (201303108).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, XD., Yu, XH., Zhou, SM. et al. De novo assembly and characterization of the Welsh onion (Allium fistulosum L.) transcriptome using Illumina technology. Mol Genet Genomics 291, 647–659 (2016). https://doi.org/10.1007/s00438-015-1131-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-015-1131-6