Abstract
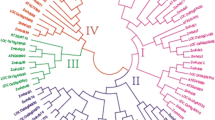
TIFY, previously known as ZIM, comprises a plant-specific family annotated as transcription factors that might play important roles in stress response. Despite TIFY proteins have been reported in Arabidopsis and rice, a comprehensive and systematic survey of ZmTIFY genes has not yet been conducted. To investigate the functions of ZmTIFY genes in this family, we isolated and characterized 30 ZmTIFY (1 TIFY, 3 ZML, and 26 JAZ) genes in an analysis of the maize (Zea mays L.) genome in this study. The 30 ZmTIFY genes were distributed over eight chromosomes. Multiple alignment and motif display results indicated that all ZmTIFY proteins share two conserved TIFY and Jas domains. Phylogenetic analysis revealed that the ZmTIFY family could be divided into two groups. Putative cis-elements, involved in abiotic stress response, phytohormones, pollen grain, and seed development, were detected in the promoters of maize TIFY genes. Microarray data showed that the ZmTIFY genes had tissue-specific expression patterns in various maize developmental stages and in response to biotic and abiotic stresses. The results indicated that ZmTIFY4, 5, 8, 26, and 28 were induced, while ZmTIFY16, 13, 24, 27, 18, and 30 were suppressed, by drought stress in the maize inbred lines Han21 and Ye478. ZmTIFY1, 19, and 28 were upregulated after infection by three pathogens, whereas ZmTIFY4, 13, 21, 23, 24, and 26 were suppressed. These results indicate that the ZmTIFY family may have vital roles in response to abiotic and biotic stresses. The data presented in this work provide vital clues for further investigating the functions of the genes in the ZmTIFY family.







Similar content being viewed by others
Abbreviations
- ABA:
-
Abscisic acid
- CCT:
-
CONSTANS, CO-like, TOC1
- JA:
-
Jasmonic acid
- JAZ:
-
JASMONATE ZIM-DOMAIN
- PPD:
-
PEAPOD
- ZIM:
-
Zinc-finger protein expressed in inflorescence meristem
References
Allen RD, Bernier F, Lessard PA, Beachy RN (1989) Nuclear factors interact with a soybean beta-conglycinin enhancer. Plant Cell 1:623–631
Aparicio-Fabre R, Guillen G, Loredo M, Arellano J, Valdes-Lopez O, Ramirez M, Iniguez LP, Panzeri D, Castiglioni B, Cremonesi P, Strozzi F, Stella A, Girard L, Sparvoli F, Hernandez G (2013) Common bean (Phaseolus vulgaris L.) PvTIFY orchestrates global changes in transcript profile response to jasmonate and phosphorus deficiency. BMC Plant Biol 13:26
Bai Y, Meng Y, Huang D, Qi Y, Chen M (2011) Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 98:128–136
Bailey TL, Boden M, Buske FA, Martin F, Grant CE, Clernenti L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucl Acids Res 37:W202–W208
Chung HS, Niu Y, Browse J, Howe GA (2009) Top hits in contemporary JAZ: an update on jasmonate signaling. Phytochemistry 70:1547–1559
Devoto A, Ellis C, Magusin A, Chang HS, Chilcott C, Zhu T, Turner JG (2005) Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol Biol 58:497–513
Ghareeb H, Becker A, Iven T, Feussner I, Schirawski J (2011) Sporisorium reilianum infection changes inflorescence and branching architectures of maize. Plant Physiol 156:2037–2052
Guo AY, Zhu QH, Chen X, Luo JC (2007) GSDS: a gene structure display server. Yi Chuan 29:1023–1026
Haberer G, Mader MT, Kosarev P, Spannagl M, Yang L, Mayer KF (2006) Large-scale cis-element detection by analysis of correlated expression and sequence conservation between Arabidopsis and Brassica oleracea. Plant Physiol 142:1589–1602
Hamilton DA, Schwarz YH, Mascarenhas JP (1998) A monocot pollen-specific promoter contains separable pollen-specific and quantitative elements. Plant Mol Biol 38:663–669
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucl Acids Res 27:297–300
Jiang Y, Deyholos MK (2006) Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol 6:25
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Major IT, Constabel CP (2006) Molecular analysis of poplar defense against herbivory: comparison of wound- and insect elicitor-induced gene expression. New Phytol 172:617–635
Merika M, Orkin SH (1993) DNA-binding specificity of GATA family transcription factors. Mol Cell Biol 13:3999–4010
Nishii A, Takemura M, Fujita H, Shikata M, Yokota A, Kohchi T (2000) Characterization of a novel gene encoding a putative single zinc-finger protein, ZIM, expressed during the reproductive phase in Arabidopsis thaliana. Biosci Biotechnol Biochem 64:1402–1409
Qin Y, Ye H, Tang N, Xiong L (2009) Systematic identification of X1-homologous genes reveals a family involved in stress responses in rice. Plant Mol Biol 71:483–496
Rao MV, Lee H, Creelman RA, Mullet JE, Davis KR (2000) Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12:1633–1646
Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66:553–563
Sekhon RS, Childs KL, Santoro N, Foster CE, Buell CR, de Leon N, Kaeppler SM (2012) Transcriptional and metabolic analysis of senescence induced by preventing pollination in maize. Plant Physiol 159:1730–1744
Shikata M, Takemura M, Yokota A, Kohchi T (2003) Arabidopsis ZIM, a plant-specific GATA factor, can function as a transcriptional activator. Biosci Biotechnol Biochem 67:2495–2497
Shikata M, Matsuda Y, Ando K, Nishii A, Takemura M, Yokota A, Kohchi T (2004) Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J Exp Bot 55:631–639
Staswick PE (2008) JAZing up jasmonate signaling. Trends Plant Sci 13:66–71
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G (2007) The tify family previously known as ZIM. Trends Plant Sci 12:239–244
Voll LM, Horst RJ, Voitsik AM, Zajic D, Samans B, Pons-Kuhnemann J, Doehlemann G, Munch S, Wahl R, Molitor A, Hofmann J, Schmiedl A, Waller F, Deising HB, Kahmann R, Kamper J, Kogel KH, Sonnewald U (2011) Common motifs in the response of cereal primary metabolism to fungal pathogens are not based on similar transcriptional reprogramming. Front Plant Sci 2:39
Wasternack C (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot 100:681–697
White DW (2006) PEAPOD regulates lamina size and curvature in Arabidopsis. Proc Natl Acad Sci USA 103:13238–13243
Ye H, Du H, Tang N, Li X, Xiong L (2009) Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol Biol 71:291–305
Zhang Y, Gao M, Singer SD, Fei Z, Wang H, Wang X (2012) Genome-wide identification and analysis of the TIFY gene family in grape. PLoS One 7:e44465
Zhang Z, Chen Y, Zhao D, Li R, Wang H, Zhang J, Wei J (2014) X1-homologous genes family as central components in biotic and abiotic stresses response in maize (Zea mays L.). Funct Integr Genomics 14:101–110
Zheng J, Fu J, Gou M, Huai J, Liu Y, Jian M, Huang Q, Guo X, Dong Z, Wang H, Wang G (2010) Genome-wide transcriptome analysis of two maize inbred lines under drought stress. Plant Mol Biol 72:407–421
Zhu D, Bai X, Chen C, Chen Q, Cai H, Li Y, Ji W, Zhai H, Lv D, Luo X, Zhu Y (2011) GsTIFY10, a novel positive regulator of plant tolerance to bicarbonate stress and a repressor of jasmonate signaling. Plant Mol Biol 77:285–297
Acknowledgments
We are grateful to our editors and reviewers for their helpful comments and the groups who submitted the microarray data to the public databases. This work was supported in part by the Beijing Nova Program (No. Z121105002512031), the National Special Program of Transgenic Research (2014ZX08003-003B), the Youth Foundation of Beijing Academy of Agriculture and Forestry Sciences (No. QNJJ201303), and the Beijing Academy of Agriculture and Forestry Sciences (No. KJCX20140202).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Lai.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, Z., Li, X., Yu, R. et al. Isolation, structural analysis, and expression characteristics of the maize TIFY gene family. Mol Genet Genomics 290, 1849–1858 (2015). https://doi.org/10.1007/s00438-015-1042-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-015-1042-6