Abstract
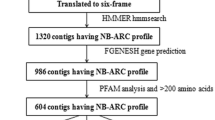
Host resistance is the most economical, effective and ecologically sustainable method of controlling diseases in crop plants. In bread wheat, despite the high number of resistance loci that have been cataloged to date, only few have been cloned, underlying the need for genomics-guided investigations capable of providing a prompt and acute knowledge on the identity of effective resistance genes that can be used in breeding programs. Proteins with a nucleotide-binding site (NBS) encoded by the major plant disease resistance (R) genes play an important role in the responses of plants to various pathogens. In this study, a comprehensive analysis of NBS-encoding genes within the whole wheat genome was performed, and the genome scale characterization of this gene family was established. From the recently published wheat genome sequence, we used a data mining and automatic prediction pipeline to identify 580 complete ORF candidate NBS-encoding genes and 1,099 partial-ORF ones. Among complete gene models, 464 were longer than 200 aa, among them 436 had less than 70 % of sequence identity to each other. This gene models set was deeply characterized. (1) First, we have analyzed domain architecture and identified, in addition to typical domain combinations, the presence of particular domains like signal peptides, zinc fingers, kinases, heavy-metal-associated and WRKY DNA-binding domains. (2) Functional and expression annotation via homology searches in protein and transcript databases, based on sufficient criteria, enabled identifying similar proteins for 60 % of the studied gene models and expression evidence for 13 % of them. (3) Shared orthologous groups were defined using NBS-domain proteins of rice and Brachypodium distachyon. (4) Finally, alignment of the 436 NBS-containing gene models to the full set of scaffolds from the IWGSC’s wheat chromosome survey sequence enabled high-stringence anchoring to chromosome arms. The distribution of the R genes was found balanced on the three wheat sub-genomes. In contrast, at chromosome scale, 50 % of members of this gene family were localized on 6 of the 21 wheat chromosomes and ~22 % of them were localized on homeologous group 7. The results of this study provide a detailed analysis of the largest family of plant disease resistance genes in allohexaploid wheat. Some structural traits reported had not been previously identified and the genome-derived data were confronted with those stored in databases outlining the functional specialization of members of this family. The large reservoir of NBS-type genes presented and discussed will, firstly, form an important framework for marker-assisted improvement of resistance in wheat, and, secondly, open up new perspectives for a better understanding of the evolution dynamics of this gene family in grass species and in polyploid systems.







Similar content being viewed by others
References
Adhikari TB, Cavaletto JR, Dubcovsky J, Gieco JO, Schlatter AR, Goodwin SB (2004) Molecular mapping of the Stb4 gene for resistance to Septoria tritici blotch in wheat. Phytopathol 94:1198–1206
Akhunov ED, Sehgal S, Liang H, Wang S, Akhunova AR, Kaur G, Li W, Forrest KL, See D, Šimková H, Ma Y, Hayden MJ, Luo M, Faris JD, Dolezel J, Gill BS (2013) Comparative analysis of syntenic genes in grass genomes reveals accelerated rates of gene structure and coding sequence evolution in polyploid wheat. Plant Physiol 161:252–265
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25:3389–3402
Ameline-Torregrosa C, Wang BB, O’Bleness MS, Deshpande S, Zhu H, Roe B, Young ND, Cannon SB (2008) Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant medicago truncatula. Plant Physiol 146:5–21
Arraiano LS, Worland AJ, Ellerbrook C, Brown JKM (2001) Chromosomal location of a gene for resistance to septoria tritici blotch (Mycosphaerella graminicola) in the hexaploid wheat ‘Synthetic 6x’. Theor Appl Genet 103:758–764
Ashfield T, Ashley NE, Pfeil BE, Chen NWG, Podicheti R, Ratnaparkhe MB, Ameline-Torregrosa C, Denny R, Cannon S, Doyle JJ, Geffroy V, Roe BA, Saghai Maroof MA, Young ND, Innes RW (2012) Evolution of a complex disease resistance gene cluster in diploid Phaseolus and tetraploid Glycine. Plant Physiol 159(1):336–354
Bella J, Hindle KL, McEwan PA, Lovell SC (2008) The leucine-rich repeat structure. Cmls-cell Mol Life Sci 277:519–527
Brenchley R, Spannagl M, Pfeifer M, Barker GLA, D’Amore R, Allen AM, McKenzie N, Kramer M, Kerhornou A, Bolser D, Kay S, Waite D, Trick M, Bancroft I, Gu Y, Huo N, Luo MC, Sehga S, Gill B, Kianian S, Anderson O, Kersey P, Dvorak J, McCombie WR, Hall A, Mayer KFX, Edwards KJ, Bevan MW, Hall N (2012) Analysis of the breadwheat genome using whole-genome shotgun sequencing. Nature 491:705–710
Bull PC, Cox DW (1994) Wilson disease and Menkes disease: new handles on heavy-metal transport. Trends Genet 10(7):246–252
Chin DB, Arroya-Garcia R, Ochoa OE, Keselli RV, Lavelle DO, Michelmore RW (2001) Recombination and spontaneous mutation at the major cluster of resistance genes in lettuce (Lactuca sativa). Genetics 157:831–849
Cloutier S, McCallum BD, Loutre C, Banks TW, Wicker T, Feuillet C, Keller B, Jordan MC (2007) Leaf rust resistance gene Lr1, isolated from bread wheat (Triticum aestivum L.) is a member of the large psr567 gene family. Plant Mol Biol 65:93–106
Deslandes L, Olivier J, Theulieres F, Hirsch J, Feng DX, Bittner-Eddy PD, Beynon J, Marco Y (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci USA 99:2404–2409
D’Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M, Da Silva C, Jabbari K, Cardi C, Poulain J, Souquet M, Labadie K, Jourda C, Lengellé J, Rodier-Goud M, Alberti A, Bernard M, Correa M, Ayyampalayam S, Mckain MR, Leebens-Mack J, Burgess D, Freeling M, Mbéguié-A-Mbéguié D, Chabannes M, Wicker T, Panaud O, Barbosa J, Hribova E, Heslop-Harrison P, Habas R, Rivallan R, Francois P, Poiron C, Kilian A, Burthia D, Jenny C, Bakry F, Brown S, Guignon V, Kema G, Dita M, Waalwijk C, Joseph S, Dievart A, Jaillon O, Leclercq J, Argout X, Lyons E, Almeida A, Jeridi M, Dolezel J, Roux N, Risterucci AM, Weissenbach J, Ruiz M, Glaszmann JC, Quétier F, Yahiaoui N, Wincker P (2012) The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488:213–217. doi:10.1038/nature11241
Ellis J, Jones D (2003) Plant disease resistance genes. In: Ezekowitz A, Hoffman J (eds) Innate immunity. Humana Press Inc., New Jersey, pp 27–44
Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10:366–371
Feuillet C, Travella S, Stein N, Albar L, Nublat A, Keller B (2003) Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc Natl Acad Sci USA 100:15253–15258
Fones H, Davis CAR, Rico A, Fang F, Smith JAC, Preston GM (2010) Metal hyperaccumulation armors plants against disease. PLoS Pathog 6(9):e1001093. doi:10.1371/journal.ppat.1001093
Friedman AR, Baker BJ (2007) The evolution of resistance genes in multiprotein plant resistance systems. Curr Opin Genet Dev 17:493–499
Glowacki S, Macioszek VK, Kononowicz AK (2011) R proteins as fundamentals of plant innate immunity. Cell Mol Biol Lett 16:1–24
Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, Hadley D, Hutchison D, Martin C, Katagiri F, Lange BM, Moughamer T, Xia Y, Budworth P, Zhong J, Miguel T, Paszkowski U, Zhang S, Colbert M, Sun WL, Chen L, Cooper B, Park S, Wood TC, Mao L, Quail P, Wing R, Dean R, Yu Y, Zharkikh A, Shen R, Sahasrabudhe S, Thomas A, Cannings R, Gutin A, Pruss D, Reid J, Tavtigian S, Mitchell J, Eldredge G, Scholl T, Miller RM, Bhatnagar S, Adey N, Rubano T, Tusneem N, Robinson R, Feldhaus J, Macalma T, Oliphant A, Briggs S (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100
Huang X, Wang L, Xu M, Röder M (2003) Microsatellite mapping of the powdery mildew resistance gene Pm5e in common wheat (Triticum aestivum L.). Theor Appl Genet 106:858–865
Huang L, Brooks S, Li W, Fellers J, Nelson JC, Gill B (2009) Evolution of new disease specificity at a simple resistance locus in a crop-weed complex: reconstitution of the Lr21 gene in wheat. Genetics 182(2):595–602
International Barley Genome Sequencing Consortium (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491:711–716
Jamali MK, Kazi TG, Arain MB, Afridi HI, Jalbani N, Kandhro GA, Shah AQ, Baig JA (2009) Heavy metal accumulation in different varieties of wheat (Triticum aestivum L.) grown in soil amended with domestic sewage sludge. J Hazardous Materials 164:1386–1391
Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, Appels R, Pfeifer M, Tao Y, Zhang X, Jing R, Zhang C, Ma Y, Gao L, Gao C, Spannagl M, Mayer KFX, Li D, Pan S, Zheng F, Hu Q, Xia X, Li J, Liang Q, Chen J, Wicker T, Gou C, Kuang H, He G, Luo Y, Keller B, Xia Q, Lu P, Wang J, Zou H, Zhang R, Xu J, Gao J, Middleton C, Quan Z, Liu G, Wang J, International Wheat Genome Sequencing Consortium, Yang H, Liu X, He Z, Mao L, Wang J (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496:91–95
Jones DA, Jones JDG (1997) The role of leucine-rich repeat proteins in plant defences. Adv Bot Res 24:90–167
Kajava AV (1998) Structural diversity of leucine-rich repeat proteins. J Mol Biol 277:519–527
Kanzaki K, Saitoh H, Fujisaki K, Kobayashi M, Ito K, Kanzaki E, Mitsuoka C, Banfield M, Kamoun S, Terauchi R (2013) Rice proteins that interact with the magnaporthe oryzae avirulence effector AVR-Pik (A-22). In: International rice blast conference, Jeju, Korea, August 20–24, 2013, Proceedings book, p 98. http://irbc2013.riceblast.snu.ac.kr/attach/IRBC2013_proceeding_book.pdf
Karatas M, Dursun S, Guler E, Ozdemir C, Argun ME (2006) Heavy metal accumulation in wheat plants irrigated by waste water. Cellulose Chem Technol 40:575–579
Kohler A, Rinaldi C, Duplessis S, Baucher M, Geelen D, Duchaussoy F, Meyers BC, Boerjan W, Martin F (2008) Genome-wide identification of NBS resistance genes in Populus trichocarpa. Plant Mol Biol 66:619–636
Krattinger SG, Lagudah ES, Wicker T, Risk JM, Ashton AR, Selter LL, Matsumoto T, Keller B (2011) Lr34 multi-pathogen resistance ABC transporter: molecular analysis of homoeologous and orthologous genes in hexaploid wheat and other grass species. Plant J 65:392–403
Leister RT, Dahlbeck D, Day B, Li Y, Chesnokova O, Staskawicz BJ (2005) Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 17:1268–1278
Li Y, Niu YC (2007) Identification of molecular markers for wheat stripe rust resistance gene Yr6. Acta Agr Boreali-Sinica 22:189–192
Lin F, Xu SC, Zhang LJ, Miao Q, Zhai Q, Li L (2005) SSR marker of wheat stripe rust resistance gene Yr2. J Tritical Crops 25:17–19
Ling HQ, Zhao S, Liu D, Wang J, Sun H, Zhang C, Fan H, Li D, Dong L, Tao Y, Gao C, Wu H, Li Y, Cui Y, Guo X, Zheng S, Wang B, Yu K, Liang Q, Yang W, Lou X, Chen J, Feng M, Jian J, Zhang X, Luo G, Jiang Y, Liu J, Wang Z, Sha Y, Zhang B, Wu H, Tang D, Shen Q, Xue P, Zou S, Wang X, Liu X, Wang F, Yang Y, An X, Dong Z, Zhang K, Zhang X, Luo MC, Dvorak J, Tong Y, Wang J, Yang H, Li Z, Wang D, Zhang A, Wang J (2013) Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496:87–90
Liu J, Coaker G (2008) Nuclear trafficking during plant innate immunity. Mol Plant 1(3):411–422
Lozano R, Ponce O, Ramirez M, Mostajo N, Orjeda G (2012) Genome-wide identification and mapping of NBS-encoding resistance genes in solanum tuberosum group Phureja. PLoS ONE 7(4):e34775. doi:10.1371/journal.pone.0034775
Marone D, Russo MA, Laidò G, De Leonardis AM, Mastrangelo AM (2013a) Plant nucleotide binding site–leucine-rich repeat (NBS–LRR) genes: active guardians in host defense responses. Int J Mol Sci 14:7302–7326
Marone D, Russo MA, Laidò G, De Vita P, Papa R, Blanco A, Gadaleta A, Rubiales D, Mastrangelo AM (2013b) Genetic basis of qualitative and quantitative resistance to powdery mildew in wheat: from consensus regions to candidate genes. BMC Genom 14:562. doi:10.1186/1471-2164-14-562
Mathé C, Sagot M-F, Schiex T, Rouzé P (2002) Current methods of gene prediction, their strengths and weaknesses. Nucl Acids Res 30:4103–4117. doi:10.1093/nar/gkf543
McIntosh R, Luig N, Baker E (1967) Genetic and cytogenetic studies of stem rust, leaf rust, and powdery mildew resistances in Hope and related wheat cultivars. Aust J Biol Sci 20:1181–1192
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotidebinding superfamily. Plant J 20:317–332
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS–LRR–encoding genes in Arabidopsis. Plant Cell Online 15:809–834
Miller RN, Bertioli DJ, Baurens FC, Santos CM, Alves PC, Martins NF, Togawa RC, Souza MT, Pappas GJ (2008) Analysis of non-TIR–NBS–LRR resistance gene analogs in Musa acuminata Colla: isolation, RFLP marker development, and physical mapping. BMC Plant Biol 8:15. doi:10.1186/1471-2229-8-15
Moffett P, Farnham G, Peart J, Baulcombe DC (2002) Interaction between domains of a plant NBS–LRR protein in disease resistance-related cell death. EMBO J 21:4511–4519
Monosi B, Wisser RJ, Pennill L, Hulbert SH (2004) Full-genome analysis of resistance gene homologues in rice. Theor Appl Genet 109:1434–1447
Mun JH, Yu HJ, Park S, Park BS (2009) Genome-wide identification of NBS encoding resistance genes in Brassica rapa. Mol Genet Genomics 282:617–631
Nematollahi G, Mohler V, Wenzel G, Zeller FJ, Hsam SLK (2008) Microsatellite mapping of powdery mildew resistance allele Pm5d from common wheat line IGV1-455. Euphytica 159:307–313
Noutoshi Y, Ito T, Seki M, Nakashita H, Yoshida S, Marco Y, Shirasu K, Shinozaki KA (2005) Single amino acid insertion in the WRKY domain of the Arabidopsis TIR–NBS–LRR–WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J 43:873–888
Oakley MG, Hollenbeck JJ (2001) The design of antiparallel coiled coils. Curr Opin Struct Biol 11:450–457
Okuyama Y, Kanzaki H, Abe A, Yoshida K, Tamiru M, Saitoh H, Fujibe T, Matsumura H, Shenton M, Clark DG, Undan J, Ito A, Sone T, Terauchi R (2011) A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS–LRR protein genes. Plant J 66:467–479
Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Mehboob-ur-Rahman, Ware D, Westhoff P, Mayer KF, Messing J, Rokhsar DS (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Periyannan S, Moore J, Ayliffe M, Bansal U, Wang X, Huang L, Deal K, Luo M, Kong X, Bariana H, Mago R, McIntosh R, Dodds P, Dvorak J, Lagudah E (2013) The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 341:786–788. doi:10.1126/science.1239028
Porter BW, Paidi M, Ming R, Alam M, Nishijima WT, Zhu YJ (2009) Genome-wide analysis of Carica papaya reveals a small NBS resistance gene family. Mol Genet Genomics 281:609–626
Rairdan GJ, Moffett P (2006) Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18:2082–2093
Saintenac C, Zhang W, Salcedo A, Rouse MN, Trick HN, Akhunov E, Dubcovsky J (2013) Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science 341:783–786. doi:10.1126/science.1239022
Salamov A, Solovyev V (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10:516–522
Sanseverino W, Ercolano MR (2012) In silico approach to predict candidate R proteins and to define their domain architecture. BMC Res Notes 5:678. doi:10.1186/1756-0500-5-678
Shang J, Tao Y, Chen X, Zou Y, Lei C, Wang J, Li X, Zhao X, Zhang M, Lu Z, Xu J, Cheng Z, Wan J, Zhu L (2009) Identification of a new rice blast resistance gene, Pid3, by genome wide comparison of paired nucleotide-binding site leucine-rich repeat genes and their pseudogene alleles between the two sequenced rice genomes. Genetics 182:1303–1311
Sharma I (2012) Diseases in wheat crops, an introduction. In: Sharma I (ed) Disease resistance in wheat. CABI, Oxfordshire, pp 1–17
Shen KA, Chin DB, Arroyo-Garcia R, Ochoa OE, Lavelle DO, Wroblewski T, Meyers BC, Michelmore RW (2002) Dm3 is one member of a large constitutively expressed family of nucleotide binding site-leucine-rich repeat encoding genes. Mol Plant-Microbe Interact 15:251–261
Spielmeyer W, McIntosh RA, Kolmer J, Lagudah ES (2005) Powdery mildew resistance and Lr34/Yr18 genes for durable resistance to leaf and stripe rust cosegregate at a locus on the short arm of chromosome 7D of wheat. Theor Appl Genet 111:731–735
Sun Q, Collins NC, Ayliffe M, Smith SM, Drake J, Pryor T, Hulbert SH (2001) Recombination between paralogues at the Rp1 rust resistance locus in maize. Genetics 158:423–438
Takken FL, Albrecht M, Tameling WI (2006) Resistance proteins: molecular switches of plant defence. Curr Opin Plant Biol 9:383–390
Tan S, Wu S (2012) Genome wide analysis of nucleotide-binding site disease resistance genes in Brachypodium distachyon. Comp Funct Genomics. doi:10.1155/2012/418208
Tian D, Traw M, Chen J, Kreitman M, Bergelson J (2003) Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423:74–77
Tosa Y, Sakai K (1990) The genetics of resistance of hexaploid wheat to the wheatgrass powdery mildew fungus. Genome 33:225–230
Traut TW (1994) The functions and consensus motifs of nine types of peptide segments that form different types of nucleotide binding sites. Eur J Biochem 222:9–19
Ulker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7:491–498
van Ooijen G, van den Burg HA, Cornelissen BJC, Takken FLW (2007) Structure and function of resistance proteins in solanaceous plants. Ann Rev Phytopathol 45:43–72
Wan H, Yuan W, Bo K, Shen J, Pang X, Chen J (2013) Genome-wide analysis of NBS-encoding disease resistance in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genom 14:109
Wang Y, Chen J-Q, Araki H, Jing Z, Jiang K, Shen J, Tian D (2004) Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol Genet Genomics 271:402–415
Weng Y, Li W, Devkota RN, Rudd JC (2005) Microsatellite markers associated with two Aegilops tauschii-derived greenbug resistance loci in wheat. Theor Appl Genet 110:462–469
Xiao M, Song F, Jiao J, Wang X, Xu H, Li H (2013) Identification of the gene Pm47 on chromosome 7BS conferring resistance to powdery mildew in the Chinese wheat landrace Hongyanglazi. Theor Appl Genet 126:1397–1403
Yahiaoui N, Srichumpa P, Dudler R, Keller B (2004) Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J 37(4):528–538
Yang S, Zhang X, Yue JX, Tian D, Chen JQ (2008) Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genomics 280:187–198
Zhou T, Wang Y, Chen JQ, Araki H, Jing Z, Jiang K, Shen J, Tian D (2004) Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR–NBS–LRR genes. Mol Genet Genomics 271:402–415
Zhu LC, Smith CM, Fritz A, Boyko E, Voothuluru P, Gill BS (2005) Inheritance and molecular mapping of new greenbug resistance genes in wheat germplasms derived from Aegilops tauschii. Theor Appl Genet 111:831–837
Acknowledgments
We gratefully acknowledge Jacques-Déric Rouault (Laboratoire Evolution, Génome et Spéciation, CNRS, Gif sur Yvette, France) for providing training; Abdelkader Aïnouche and Malika-Lily Aïnouche (UMR-CNRS Ecobio, Université de Rennes-1, France) for useful discussion and manuscript revision, and Rafika Challouf (Institut National des Sciences et Technologies de la Mer, INSTM, Monastir, Tunisia) for providing help in computational analyses. This study was financially supported by the Tunisian Ministry of Higher Education and Scientific Research.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bouktila, D., Khalfallah, Y., Habachi-Houimli, Y. et al. Full-genome identification and characterization of NBS-encoding disease resistance genes in wheat. Mol Genet Genomics 290, 257–271 (2015). https://doi.org/10.1007/s00438-014-0909-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-014-0909-2