Abstract
In this paper, we describe a novel processing strategy for the spatial normalization of ultrahigh resolution magnetic resonance imaging (MRI) data of small ex vivo samples into MNI standard space. We present a multistage scanning and registration method for data of the subthalamic nucleus (STN) obtained using ultrahigh 7 T MRI on four human postmortem brain samples. Four whole brains were obtained and subjected to multistage MRI scanning, corresponding to four different brain dissection stages. Data sets were acquired with an isotropic resolution of 100 μm enabling accurate manual segmentation of the STN. Spatial normalization to MNI reference space was performed, probability maps were calculated, and results were cross-checked with an independent in vivo dataset showing significant overlay. Normalization of results obtained from small tissue samples into MNI standard space will facilitate comparison between individual subjects, as well as between studies. When combining ultrahigh resolution MRI of ex vivo samples with histological studies via blockface imaging, our method enables further insight and inference as multimodal data can be compared within the same reference space. This novel technique may be of value for research purposes using functional MRI techniques, and in the future may be of assistance for anatomical orientation in clinical practice.
Similar content being viewed by others
Introduction
Recent studies have recognized the importance of the integration of data available from histological analyses with noninvasive imaging techniques (Annese 2012; Yang et al. 2013). Histological analyses have been used to provide impressive three-dimensional (3D) reconstructions of the human brain (Amunts et al. 2013). These studies are valuable, but the techniques used may not be universally suitable for routine implementation (Annese 2012). This is illustrated by the low number of observations described in these studies, n = 1, 1 and 2 respectively (Annese 2012; Amunts et al. 2013; Yang et al. 2013). One of the limitations that neuroscientists working with human postmortem brain material often encounter is the availability of only small samples of brain tissue. The need for methods allowing the translation of histological data from small tissue samples into standard MNI space is therefore evident. Here, we describe a method allowing detailed histological analyses of smaller tissue blocks, and the translation of the data into MNI space and information relevant for the in vivo situation. The applicability of the created pipeline is illustrated using the human subthalamic nucleus (STN).
The STN, a small nucleus in the basal ganglia, plays a crucial role in the regulation of motor, limbic, and associative functions (Temel et al. 2005). Visualization of the STN in postmortem tissue samples using histological stainings allows for a reliable determination of the STN's location as well as its anatomical borders. The STN is located as a roof above the substantia nigra (SN) pars compacta, the caudal part of the pars reticulata, and the comb system, caudal to the red nucleus. Discerning the borders of the STN becomes challenging when the STN is visualized using noninvasive techniques, such as magnetic resonance imaging (MRI). The STN's precise anatomical delineation on 1.5 and 3 T MRI is difficult to determine in view of the nucleus’ small volume, and its heterogeneous iron content (de Hollander et al. 2014). The border between the STN and the more medially located iron-rich SN is not visible using conventional MRI. These limitations hamper (functional) MRI research on STN function, since manual segmentation is challenging on 1.5 and 3 T MRI, and the subsequent registration of the STN in common space is not trivial. An alternative approach for STN segmentation on MRI is therefore warranted.
Today ultrahigh (UH) resolution MRI is available for research purposes and recent studies have shown that the STN can be visualized with greater detail using 7 T or even 9.4 T MRI (Cho et al. 2010; Forstmann et al. 2010, 2012; Massey et al. 2012; Schafer et al. 2012). Unfortunately, the lengthy scanning sequences used for visualization using UH resolution MRI of the STN are not suitable for functional research or diagnostic purposes, and high magnetic field scanners are not widely available. In addition, UH resolution scans are highly susceptible to motion artifacts, which is particularly relevant for scanning in patients suffering from movement disorders. Recent developments in prospective motion correction methods are showing great promise in vivo (Schulz et al. 2012). UH resolution MRI has great value for detailed studies on postmortem human brain specimens. Massey et al. (2012) recently demonstrated the anatomical delineation of the STN in human postmortem brain material using UH resolution 9.4 T MRI, which was confirmed by histological staining of the tissues.
Human brain specimens are not readily available and samples are generally small, and do not always contain anatomical landmarks required for registration of the images into standard space. The absence of measurements relative to the midpoint between the mamillothalamic tract and the fornix renders it impossible to register findings into standard space. In addition, available tools for spatial normalization of MR data are generally developed for whole brain scans. An alternative registration approach not requiring these measurements would, therefore, be of great value to the scientific community.
Structures surrounding the STN, such as the SN and red nucleus can more easily be visualized on MRI than the STN. The SN and red nucleus contain large quantities of iron, which presents as a hypointense signal on T2 and T2*-weighted images, and high susceptibility values in susceptibility maps (Dormont et al. 2004; Vertinsky et al. 2009; Schafer et al. 2012). These and other landmarks are important for anatomical orientation when the STN itself is difficult to visualize, and these structures could be utilized in the creation of probability maps for STN localization in MNI common space. Such probability maps could then ultimately be used by neuroscientists for localization of the STN, and in the future for neurosurgeons as an additional tool to assist anatomical orientation in preparation for stereotactic-guided DBS surgery.
In the present study, we visualized the human STN in postmortem tissue samples, as demonstrated by Massey et al. (2012) using UH resolution MRI. The aim of the present study was to develop a multistage registration approach using linear registration and nonlinear warping allowing us to normalize microstructural postmortem STN data into a common reference system, and to assess its applicability to the in vivo situation. We tested the validity of our approach by comparing the ex vivo data to our earlier in vivo observations in an age-appropriate control group (Keuken et al. 2013). In addition, corresponding scans of the intact postmortem brains were used for comparison. The developed method will allow us and other researchers to transfer results obtained from small brain samples, including histological and MR microscopy results back into common space. We created an initial STN probability map with an unprecedented 0.4-mm isotropic resolution. Finally, we overlaid this template on a dataset that we published earlier for comparison, and to determine whether the location and volume of the STN of the postmortem samples matched with that described in their appropriate in vivo age group (middle aged or elderly) (Keuken et al. 2013).
Materials and methods
Postmortem brain specimens
Four postmortem human brains were obtained in accordance with all legal requirements. Tissues were fixed in 4 or 10 % formalin. During the fixation process, brain specimens were suspended from the basal arteries to prevent anatomical deformation. Fixative was changed regularly to ensure optimal preservation of the tissue. Clinicopathological data are presented in Table 1.
Tissue analysis
Prior to further processing, a whole brain scan was obtained using MR (stage 1). The STN area was then further dissected through three cutting stages (stages 2–4). Tissue blocks cut at individual stages were scanned using MRI (Fig. 1). All blocks were cut perpendicular to the anterior commissure–posterior commissure (AC–PC) line.
MR image acquisition
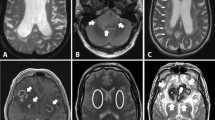
All brains were scanned in four consecutive stages. Scan parameters are presented in Table 2. After an initial whole brain scan (stage 1), brainstem and cerebellum were removed at the level of the cerebral peduncle and hemispheres were separated. All cutting was done perpendicular to the anterior commissure-posterior commissure (AC–PC) line and the medial wall. Coronal slabs (stage 2) were cut further with the anterior border located 2 cm anterior of AC. Slabs were 7 cm thick. For stage 3, slabs were trimmed to blocks using AC and PC as rostral and caudal borders. The superior border was cut 1 cm above the AC–PC line. The final STN stage (4) was created by cutting a cube of 2 × 2 × 2 cm with its anterior border 5 mm posterior of the AC, the inferior border was formed by the basis of the brain, and the lateral border 2 cm from the medial ventricular wall. Stage 4 blocks contained the entire STN, but not the medial-basal part of the substantia nigra, nor the most caudal part of the red nucleus. Figure 1 illustrates the four stages showing photographs and illustrative MRI data at each stage.
Magnetic resonance imaging was performed using a 7 T Siemens whole-body MR scanner with a 24-channel Nova phased-array head coil for scanning stages 1 and 2. For stages 3 and 4, we used a custom-built 120-mm square single-channel coil (subjects #04–10 and #05–10), or a 80-mm dual loop miniCP coil (subjects #01–11 and #06–12). During scanning, brain samples were mounted in acrylic spheres filled with Fomblin (Solvay Solexis) to minimize background signal. For stage 1, scans of cases #01–11 and #06–12 we used a dedicated whole brain container. This container was designed for supine scans of whole brain specimens using standard in vivo MRI coils.
T1 and T2*w scans were acquired including quantitative parametric maps using a MP2RAGE sequence (Marques et al. 2010) and a GRE FLASH sequence. MRI parameters were determined based on prior experience and additional test scans including quantitative T1 and T2* maps. MRI parameters for all acquisitions relevant for this study are summarized in Table 2. Our research group, as well as other, has successfully applied ultrahigh resolution MRI to visualize the STN in humans (Cho et al. 2010; Forstmann et al. 2010, 2012; Keuken et al. 2013; Massey et al. 2012). Accurate segmentation of the STN requires manual outlining of the nucleus based on visual inspection. Therefore, good quality contrast levels in this inhomogenous nucleus are invaluable to obtain reliable results. To this end we, and others, have previously successfully applied fast low angle shot (FLASH) MRI imaging (Cho et al. 2010; Forstmann et al. 2010, 2012; Keuken et al. 2013; Massey et al. 2012). On T2 and T2* images, the STN appears as a hypointense structure (Dormont et al. 2004). In addition, the use of FLASH allowed us to obtain high resolution images, which would not have been possible using MP2RAGE, due to the inherent limitations of the inversion times. Since T2* imaging is sensitive to iron, as well as the orientation of white matter bundles, this technique is well suited for imaging of closely connected brain areas surrounded by iron-rich structures, such as the STN.
No corrections were needed to correct for image distortions caused by susceptibility gradients. These distortions in our experience usually occur at air–tissue interfaces. In our approach, we use a sealed container filled with Fomblin. Air–tissue interfaces are therefore not present, and the associated artifacts are prevented.
STN segmentation
Subthalamic nuclei were manually outlined using FSLview in individual space by three independent researchers (AA, MCK, and CMA) using the 100-μm isotropic GRE data obtained in stage 4. Dice coefficients (Dice, 1945) were calculated to assess inter-rater variation. In one subject, the Dice coefficient was low (#05–10, dice <0.7). In this case, researchers (AA, MCK, and CMA) discussed the anatomy until consensus was reached. Mean dice coefficient over the three raters was 0.78. Segmentation data of all three raters were combined and voxels only scored by one of the raters were discarded prior to volume calculations.
Image registration and spatial normalization
Registration was performed separately for each hemisphere and in reversed order. We aligned the data of one stage to the appropriate data of the next smaller stage (stage 3–4, stage 2–3, and stage 1–2). Using a final, linear registration step, we brought all the right hemisphere data sets into alignment with the left side ones. Spatial normalization was performed using MP2RAGE T1-maps, and resampling of the data to allow comparison to the MNI template.
Linear registration was performed using SPM8 Coregister (Ashburner and Friston 2005). High resolution FLASH data were used for intraindividual linear registration across the four stages. Manual pre-alignment was applied prior to each registration step in view of the different global orientations resulting from the acquisition setups for the four MRI scanning stages. No within stage registration was performed. We registered the two hemispheric brain data sets, and flipping all data from the right hemisphere across the midsagittal plane. Data sets from all scanning stages were aligned in the native space of the individual left stage 4 scans. No re-sampling was performed throughout the entire linear registration pipeline.
Subsequent spatial normalization to MNI standard reference space was performed using the whole brain MP2RAGE T1 maps since those are more similar to the MNI template data than the GRE scans. Again manual alignment of the entire multistage data set towards the MNI template was performed followed by whole brain normalization warp using SPMs Unified Segmentation approach (Ashburner and Friston 2005). The resulting warp field allowed nonlinear normalization of data from every single stage to MNI standard reference space. Finally, we re-sampled the whole brain FLASH data at 500-μm isotropic resolution and compared the accuracy of the normalization warp with the 500-μm MNI template.
We further normalized the ultrahigh resolution stage 4 data including the STN masks. These data were re-sampled into the MNI subspace (X 5 to −30; Y 30 to −30; Z 10 to −20) at their original 100 μm resolution using nearest neighbor interpolation. Masks were combined for each hemisphere, and voxel scores were divided by the maximum number of positive ratings (4 × 3 = 12). This resulted in a normalized probability map of STNs in each hemisphere, with values between 0 and 1, a value of 1 reflecting the presence of the STN in this voxel in all four brains as rated by all three raters. These maps were resampled to 0.4-mm isotropic resolution in full-MNI space and were overlaid with the age-appropriate independent in vivo dataset for comparison (Forstmann et al. 2012; Keuken et al. 2013).
Template building
To enable the spatial normalization of small tissue samples when whole brains are not accessible and multistage scanning is therefore not possible; we used our spatially normalized ultrahigh resolution stage 4 FLASH data to create a spatial template. Right hemisphere data were flipped to the left to avoid lateralization bias. An initial template was created performing a landmark-based registration. Five landmarks identifiable in all samples were selected and the coordinates of these landmarks were averaged across the eight samples to calculate target coordinates. All data sets were transformed using a thin-plate-spline-based landmark registration (Bookstein 1989), bias field correction was performed using the N4 algorithm, and the start template was created by voxel-wise averaging.
The final template was created utilizing the buildtemplateparallel.sh script provided by the ANTS software package (Avants et al. 2011). Starting with the landmark-based pre-aligned versions of the eight individual, normalized stage 4 FLASH data sets, and the corresponding start template; every iteration was based on the resulting template and the deformed individual data of the previous iteration. A total of fifteen nonlinear registration iterations for each of the individual data sets, as well as the corresponding intermediate templates were carried out to create a final spatial template for the STN samples. We applied Greedy SyN registration with mutual information similarity measurements and set parameters according to the suggestions by Avants et al. (2011). More concretely, the gradient step size was set at 0.1, multiresolution parameters were 45 × 15 × 5 × 0 × 0, and regularization to Gauss [3,1] was performed.
Results
Using 7 T MRI, we were able to visualize the human STN borders with a high precision (Fig. 1). The anatomical detail that was obtained provides us with information on the location and volume of the STN in human postmortem brain samples (Fig. 1). The scan quality allowed manual segmentation of the STN on the FLASH images including the SN border of the STN. Anatomical detail is reflected in the Dice coefficients (mean ± SD, 0.78 ± 0.06). Note that the Dice coefficient reported here is calculated as a mean between three raters. The high anatomical detail is likely to have also contributed to the differences observed in STN volume (109 ± 46 mm3), which we reported earlier (range 52–77 mm3) (Keuken et al. 2013). The STN volume observed here was significantly larger than the STN volume in the elderly group published previously (Wilcoxon’s rank sum test with continuity correction, W = 121, p = 0.007) (Keuken et al. 2013).
We subsequently performed linear registration of the scans across the four scanning stages, which allowed us to normalize the data to MNI space utilizing a subsequent whole brain warp as part of the unified segmentation approach (Ashburner and Friston 2005). Accurate alignment across stages was achieved using manual prealignment and linear registration (Fig. 2). This method sufficiently corrected artifacts introduced by cutting or due to additional shrinkage because of continued fixation of later cutting stages. Spatial normalization was successfully performed, and is illustrated in the 500 μm MNI template shows created (Fig. 3).
Based on this information, we were able to create probability maps for STN location based on the four studied brains. These maps corresponded with the expected location of the STN within MNI standard space (Fig. 2). For comparison, we also overlaid the probability maps with independent age-appropriate in vivo datasets that we published earlier (Keuken et al. 2013) (Fig. 2). Visual inspection showed significant overlap between the in vivo and ex vivo STN masks, although the ex vivo masks had a larger volume. The results of our normalization procedure allowed us to create a spatial template (Fig. 4) that can be applied to STN tissues samples of similar size as our step 4 samples. Our template is in accordance with the group average in size shape and intensity distribution for the STN as based on the T2*-weighted input images.
Discussion
The high Dice coefficients (mean ± SD, 0.78 ± 0.06), we obtained in the present studies were higher than those described by our group for in vivo data (Keuken et al. 2013). In view of the high anatomical resolution and excellent contrast of the MRI images, it is likely that this difference is, at least partly, due to the excellent quality of the brain images. Here we report larger STN volumes (109 ± 46 mm3) as compared to our earlier findings (range 52–77 mm3) (Keuken et al. 2013). In both studies, volumes were based on the conjunction of the masks of the individual raters. The smaller STN volume, which we reported previously, can therefore most likely be attributed to conservative masking of the STN at the borders, as well as partial volume effects. Interestingly, although the number of observations we describe in these studies are limited, we find a substantial overlay of the STN location in postmortem tissues with our earlier in vivo data. This overlap supports the validity of our findings. We would like to emphasize that caution is warranted since the maps were based on a limited number of only four observations per hemisphere. The present studies were primarily aimed at developing a novel approach to translate data from small tissue samples into MNI space, not at creating a template for outlining the STN, which can be readily used for research purposes. The number of observations needs to be increased to create a more reliable template. In addition, sequence parameters as used for the STN may not yield optimal results for other brain areas, such as individual hypothalamic or cerebellar nuclei. The spatial normalization method that we describe is fully automated and can be used for other subcortical nuclei, as well as for cortical areas. Our method can also be applied for research purposes to allow for determination of STN location in functional MRI studies in healthy subjects. This comparison can be obtained using the unified segmentation approach. In addition, it could potentially serve as an atlas tool in aiding to determine STN location as a surgical target for DBS. This, however, lies beyond the scope of the present studies.
Several factors may have influenced our results. Working with human postmortem brain tissue comes with inherent limitations. Tissues are obtained with various intervals between time of death and autopsy. In addition, subjects die of various causes after variable antemortem disease length and severity. It is likely that subjects received pharmacological treatment even though these data were not available. Postmortem tissue is generally obtained from older subjects. We know from our earlier studies that age-related changes are present in STN location (Keuken et al. 2013). Three brain specimens showed comparable results. Registration of one specimen (#06–12) was found to be more challenging due to an old cerebral infarction. We managed to successfully register this brain despite the presence of a space-occupying process. Whether and to what extent the additional factors mentioned here have affected our MRI scans is unknown, but we cannot exclude it. Processing of the tissues after autopsy may have affected our results. Postmortem tissue blocks undergo fixation procedures, which may cause deformation of the tissue, including shrinkage. In addition, prolonged fixation affects the magnetic properties of the tissue (Shepherd et al. 2009). Our normalization approach takes into account tissue shrinkage, and we optimized our MRI parameters based on quantitative maps of relaxation parameters (T1/T2*). The spatial template will enable us to normalize small tissue samples in future studies. This is particularly relevant if no whole brain specimens are available, and therefore multistage scanning and registration is not possible.
This study provides valuable information on STN neuroanatomy and it has been shown in the past that postmortem observations are relevant to premortem observations, such as serum hormone concentrations as well as anthropometric parameters (Fliers et al. 1997; Alkemade et al. 2012a, b). Because human brain tissues are not readily available, often brains are divided among researchers, and small brain samples are available for individual studies. Applying the techniques described here renders it possible to process small postmortem brain specimens further, allowing detailed analysis, which can ultimately be registered back into MNI space without scanning whole brain specimens. This will provide us, for the first time, with detailed information of the STN. This technique opens a window to combine MRI and histological data into an integrated brain atlas. It also offers an opportunity to recombine observations from separate tissue blocks of the same subjects into a single analysis, as well as a comparison with future studies that can be registered into MNI common space.
We have optimized the technique for STN research in view of its important role in the basal ganglia, and as a target for deep brain stimulation in Parkinson’s disease. This technique can be used as well to process other brain structures that can be visualized using (ultrahigh resolution) MRI. Such studies will not only improve understanding of human brain anatomy, but may also facilitate anatomical orientation in clinical practice.
References
Alkemade A, Unmehopa UA, Hessel EV, Swaab DF, Kalsbeek A, Fliers E (2012a) Suppressor of cytokine signaling 3 in the human hypothalamus. Peptides 35(1):139–142. doi:10.1016/j.peptides.2012.03.004
Alkemade A, Yi CX, Pei L, Harakalova M, Swaab DF, la Fleur SE, Fliers E, Kalsbeek A (2012b) AgRP and NPY expression in the human hypothalamic infundibular nucleus correlate with body mass index, whereas changes in alphaMSH are related to type 2 diabetes. J Clin Endocrinol Metab 97(6):E925–E933. doi:10.1210/jc.2011-3259
Amunts K, Lepage C, Borgeat L, Mohlberg H, Dickscheid T, Rousseau ME, Bludau S, Bazin PL, Lewis LB, Oros-Peusquens AM, Shah NJ, Lippert T, Zilles K, Evans AC (2013) BigBrain: an ultrahigh-resolution 3D human brain model. Science 340(6139):1472–1475. doi:10.1126/science.1235381
Annese J (2012) The importance of combining MRI and large-scale digital histology in neuroimaging studies of brain connectivity and disease. Front Neuroinform 6:13. doi:10.3389/fninf.2012.00013
Ashburner J, Friston KJ (2005) Unified segmentation. NeuroImage 26(3):839–851. doi:10.1016/j.neuroimage.2005.02.018
Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC (2011) A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage 54(3):2033–2044. doi:10.1016/j.neuroimage.2010.09.025
Bookstein FL (1989) Principal Warps: thin-plate splines and the decompositions of deformations. IEEE Trans Pattern Anal Mach Intell 11(6):567–585
Cho ZH, Min HK, Oh SH, Han JY, Park CW, Chi JG, Kim YB, Paek SH, Lozano AM, Lee KH (2010) Direct visualization of deep brain stimulation targets in Parkinson disease with the use of 7-tesla magnetic resonance imaging. J Neurosurg 113(3):639–647. doi:10.3171/2010.3.JNS091385
de Hollander G, Keuken MC, Bazin PL, Weiss M, Neumann J, Reimann K, Wähnert, Turner R, Forstmann BU, Scshäfer A (2014) A gradual increase of iron toward the medial-inferior tip of the subthalamic nucleus. Hum Brain Mapp doi:10.1002/hbm.22485
Dormont D, Ricciardi KG, Tande D, Parain K, Menuel C, Galanaud D, Navarro S, Cornu P, Agid Y, Yelnik J (2004) Is the subthalamic nucleus hypointense on T2-weighted images? A correlation study using MR imaging and stereotactic atlas data. AJNR Am J Neuroradiol 25(9):1516–1523
Fliers E, Guldenaar SE, Wiersinga WM, Swaab DF (1997) Decreased hypothalamic thyrotropin-releasing hormone gene expression in patients with nonthyroidal illness. J Clin Endocrinol Metab 82(12):4032–4036
Forstmann BU, Anwander A, Schafer A, Neumann J, Brown S, Wagenmakers EJ, Bogacz R, Turner R (2010) Cortico-striatal connections predict control over speed and accuracy in perceptual decision making. Proc Natl Acad Sci USA 107(36):15916–15920. doi:10.1073/pnas.1004932107
Forstmann BU, Keuken MC, Jahfari S, Bazin PL, Neumann J, Schafer A, Anwander A, Turner R (2012) Cortico-subthalamic white matter tract strength predicts interindividual efficacy in stopping a motor response. NeuroImage 60(1):370–375. doi:10.1016/j.neuroimage.2011.12.044
Keuken MC, Bazin PL, Schafer A, Neumann J, Turner R, Forstmann BU (2013) Ultra-high 7T MRI of structural age-related changes of the subthalamic nucleus. J Neurosci 33(11):4896–4900. doi:10.1523/JNEUROSCI.3241-12.2013
Massey LA, Miranda MA, Zrinzo L, Al-Helli O, Parkes HG, Thornton JS, So PW, White MJ, Mancini L, Strand C, Holton JL, Hariz MI, Lees AJ, Revesz T, Yousry TA (2012) High resolution MR anatomy of the subthalamic nucleus: imaging at 9.4 T with histological validation. NeuroImage 59(3):2035–2044. doi:10.1016/j.neuroimage.2011.10.016
Marques JP, Kober T, Krueger G, Van der Zwaag W, Van de Moortele PF, Greutter R (2010) MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. NeuroImage 49(2):1271–1281. doi:10.1016/j.neuroimage.2009.10.002
Schafer A, Forstmann BU, Neumann J, Wharton S, Mietke A, Bowtell R, Turner R (2012) Direct visualization of the subthalamic nucleus and its iron distribution using high-resolution susceptibility mapping. Hum Brain Mapp 33(12):2831–2842. doi:10.1002/hbm.21404
Schulz J, Siegert T, Reimer E, Labadie C, Maclaren J, Herbst M, Zaitsev M, Turner R (2012) An embedded optical tracking system for motion-corrected magnetic resonance imaging at 7T. MAGMA 25(6):443–453. doi:10.1007/s10334-012-0320-0
Shepherd TM, Thelwall PE, Stanisz GJ, Blackband SJ (2009) Aldehyde fixative solutions alter the water relaxation and diffusion properties of nervous tissue. Magn Reson Med 62(1):26–34. doi:10.1002/mrm.21977
Temel Y, Blokland A, Steinbusch HW, Visser-Vandewalle V (2005) The functional role of the subthalamic nucleus in cognitive and limbic circuits. Prog Neurobiol 76(6):393–413. doi:10.1016/j.pneurobio.2005.09.005
Vertinsky AT, Coenen VA, Lang DJ, Kolind S, Honey CR, Li D, Rauscher A (2009) Localization of the subthalamic nucleus: optimization with susceptibility-weighted phase MR imaging. AJNR Am J Neuroradiol 30(9):1717–1724. doi:10.3174/ajnr.A1669
Yang S, Yang Z, Fischer K, Zhong K, Stadler J, Godenschweger F, Steiner J, Heinze HJ, Bernstein HG, Bogerts B, Mawrin C, Reutens DC, Speck O, Walter M (2013) Integration of ultra-high field MRI and histology for connectome based research of brain disorders. Front Neuroanat 7:31. doi:10.3389/fnana.2013.00031
Acknowledgments
This work was supported by The Netherlands Organization for Scientific Research (Vidi-grant, BUF), the European Research Council (BUF), and the Dutch Brain Foundation (AA and BUF). All data and analytical procedures are available upon request via the corresponding author. STN probability maps are freely available on http://www.nitrc.org/projects/atag/.
Author information
Authors and Affiliations
Corresponding author
Additional information
M. Weiss and A. Alkemade contributed equally to this work.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Weiss, M., Alkemade, A., Keuken, M.C. et al. Spatial normalization of ultrahigh resolution 7 T magnetic resonance imaging data of the postmortem human subthalamic nucleus: a multistage approach. Brain Struct Funct 220, 1695–1703 (2015). https://doi.org/10.1007/s00429-014-0754-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00429-014-0754-4