Abstract
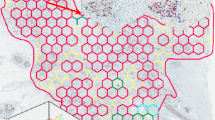
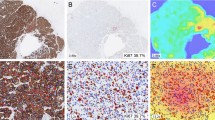
Proliferative activity, assessed by Ki67 immunohistochemistry (IHC), is an established prognostic and predictive biomarker of breast cancer (BC). However, it remains under-utilized due to lack of standardized robust measurement methodologies and significant intratumor heterogeneity of expression. A recently proposed methodology for IHC biomarker assessment in whole slide images (WSI), based on systematic subsampling of tissue information extracted by digital image analysis (DIA) into hexagonal tiling arrays, enables computation of a comprehensive set of Ki67 indicators, including intratumor variability. In this study, the tiling methodology was applied to assess Ki67 expression in WSI of 152 surgically removed Ki67-stained (on full-face sections) BC specimens and to test which, if any, Ki67 indicators can predict overall survival (OS). Visual Ki67 IHC estimates and conventional clinico-pathologic parameters were also included in the study. Analysis revealed linearly independent intrinsic factors of the Ki67 IHC variance: proliferation (level of expression), disordered texture (entropy), tumor size and Nottingham Prognostic Index, bimodality, and correlation. All visual and DIA-generated indicators of the level of Ki67 expression provided significant cutoff values as single predictors of OS. However, only bimodality indicators (Ashman’s D, in particular) were independent predictors of OS in the context of hormone receptor and HER2 status. From this, we conclude that spatial heterogeneity of proliferative tumor activity, measured by DIA of Ki67 IHC expression and analyzed by the hexagonal tiling approach, can serve as an independent prognostic indicator of OS in BC patients that outperforms the prognostic power of the level of proliferative activity.



Similar content being viewed by others
References
Esposito A, Criscitiello C, Curigliano G (2015) Highlights from the 14(th) St Gallen International Breast Cancer Conference 2015 in Vienna: Dealing with classification, prognostication, and prediction refinement to personalize the treatment of patients with early breast cancer Ecancermedicalscience 9:518. doi:10.3332/ecancer.2015.518
Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, Perou CM, Ellis MJ, Nielsen TO (2009) Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 101:736–750. doi:10.1093/jnci/djp082
Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, HJ S (2013) Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer. Ann Oncol 24:2206–2223. doi:10.1093/annonc/mdt303
Petrelli F, Viale G, Cabiddu M, Barni S (2015) Prognostic value of different cut-off levels of Ki-67 in breast cancer: a systematic review and meta-analysis of 64,196 patients. Breast Cancer Res Treat 153:477–491. doi:10.1007/s10549-015-3559-0
Lu H, Papathomas TG, van Zessen D, Palli I, de Krijger RR, van der Spek PJ, Dinjens W, Stubbs AP (2014) Automated Selection of Hotspots (ASH): enhanced automated segmentation and adaptive step finding for Ki67 hotspot detection in adrenal cortical cancer Diagn Pathol 9:216. doi:10.1186/s13000–014-0216-6
Romero Q, Bendahl PO, Ferno M, Grabau D, Borgquist S (2014) A novel model for Ki67 assessment in breast cancer. Diagn Pathol 9:118. doi:10.1186/1746-1596-9-118
Christgen M, von Ahsen S, Christgen H, Länger F, Kreipe H (2015) The region of interest (ROI) size impacts on Ki67 quantification by computer-assisted image analysis in breast cancer Human Pathology
Plancoulaine B, Laurinaviciene A, Herlin P, Besusparis J, Meskauskas R, Baltrusaityte I, Iqbal Y, Laurinavicius A (2015) A methodology for comprehensive breast cancer Ki67 labeling index with intra-tumor heterogeneity appraisal based on hexagonal tiling of digital image analysis data Virchows Arch. doi:10.1007/s00428-015-1865-x
Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, IO E (2005) High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cdna expression analyses. Int J Cancer 116:340–350. doi:10.1002/ijc.21004
Rakha EA, El-Sayed ME, Lee AHS, Elston CW, Grainge MJ, Hodi Z, Blamey RW, IO E (2008) Prognostic significance of Nottingham histologic grade in invasive breast carcinoma. Journal of Clinical Oncology 26:3153–3158. doi:10.1200/Jco.2007.15.5986
Galea MH, Blamey RW, Elston CE, IO E (1992) The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res Treat 22:207–219
Aleskandarany MA, Green AR, Rakha EA, Mohammed RA, Elsheikh SE, Powe DG, Paish EC, Macmillan RD, Chan S, Ahmed SI, IO E (2010) Growth fraction as a predictor of response to chemotherapy in node-negative breast cancer. International Journal of Cancer 126:1761–1769. doi:10.1002/ijc.24860
Barros FF, Abdel-Fatah TM, Moseley P, Nolan CC, Durham AC, Rakha EA, Chan S, Ellis IO, AR G (2014) Characterisation of HER heterodimers in breast cancer using in situ proximity ligation assay. Breast Cancer Res Treat 144:273–285. doi:10.1007/s10549-014-2871-4
Haralick RM, Shanmugan K, Distein I (1973) Textural features for image classification IEEE transactions on systems. Man, and Cybernetics SMC-3:610–621
Xuan GR, Zhang W, Chai PQ (2001) EM algorithms of Gaussian Mixture Model and Hidden Markov Model Ieee Image Proc:145–148
Dempster A, Land NM, Rubin DB (1977) Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society 39:1–38
Laurinavicius A, Laurinaviciene A, Ostapenko V, Dasevicius D, Jarmalaite S, Lazutka J (2012) Immunohistochemistry profiles of breast ductal carcinoma: factor analysis of digital image analysis data. Diagn Pathol 7:27. doi:10.1186/1746-1596-7-27
Laurinavicius A, Green AR, Laurinaviciene A, Smailyte G, Ostapenko V, Meskauskas R, Ellis IO (2015) Ki67/SATB1 ratio is an independent prognostic factor of overall survival in patients with early hormone receptor-positive invasive ductal breast carcinoma Oncotarget. doi: 10.18632/oncotarget.5838
Budczies J, Klauschen F, Sinn BV, Gyorffy B, Schmitt WD, Darb-Esfahani S, Denkert C (2012) Cutoff finder: a comprehensive and straightforward web application enabling rapid biomarker cutoff optimization. PLoS One 7:e51862. doi:10.1371/journal.pone.0051862
Laurinavicius A, Plancoulaine B, Laurinaviciene A, Herlin P, Meskauskas R, Baltrusaityte I, Besusparis J, Dasevicius D, Elie N, Iqbal Y, Bor C, Ellis IO (2014) A methodology to ensure and improve accuracy of Ki67 labelling index estimation by automated digital image analysis in breast cancer tissue Breast Cancer Res 16:R35. doi:10.1186/bcr3639
Her I (1995) Geometric transformations on the hexagonal grid. IEEE Trans Image Process 4:1213–1222. doi:10.1109/83.413166
Potts SJ, Krueger JS, Landis ND, Eberhard DA, Young GD, Schmechel SC, Lange H (2012) Evaluating tumor heterogeneity in immunohistochemistry-stained breast cancer tissue Lab Invest 92:1342–1357. doi:10.1038/labinvest.2012.91
Faratian D, Christiansen J, Gustavson M, Jones C, Scott C, Um I, Harrison DJ (2011) Heterogeneity mapping of protein expression in tumors using quantitative immunofluorescence J Vis Exp:e3334. doi:10.3791/3334
Dodd LG, Kerns BJ, Dodge RK, Layfield LJ (1997) Intratumoral heterogeneity in primary breast carcinoma: study of concurrent parameters. J Surg Oncol 64:280–287 discussion 287-288
Brown JR, DiGiovanna MP, Killelea B, Lannin DR, Rimm DL (2014) Quantitative assessment Ki-67 score for prediction of response to neoadjuvant chemotherapy in breast cancer. Lab Investig 94:98–106. doi:10.1038/labinvest.2013.128
Acknowledgments
This research is funded by the European Social Fund under the Global Grant measure, Grant #VP1-3.1-SMM-07-K-03-051.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Laurinavicius, A., Plancoulaine, B., Rasmusson, A. et al. Bimodality of intratumor Ki67 expression is an independent prognostic factor of overall survival in patients with invasive breast carcinoma. Virchows Arch 468, 493–502 (2016). https://doi.org/10.1007/s00428-016-1907-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-016-1907-z