Abstract
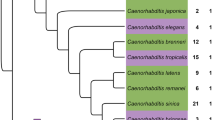
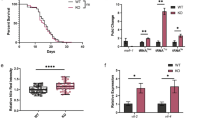
Mutations of the puromycin-sensitive aminopeptidase (Psa) orthologs of flies, mice, and plants result in meiotic errors and reduced embryonic viability. Genetic lesions of the Caenorhabditis elegans ortholog of Psa, pam-1, similarly result in dramatic reductions of worm fecundity. The gonads of animals harboring mutant pam-1 alleles display expanded populations of pachytene germinal nuclei and delayed nucleolar disassembly in the developing oocytes, phenotypes that ultimately hinder embryonic viability and overall brood sizes. PAM-1 is a member of the M1 aminopeptidase family and shares a high amount of homology with its M1 paralogs. Comparative analysis of the M1 aminopeptidase family reveals that only nine (including PAM-1) of the 17 annotated M1 aminopeptidases are predicted to be catalytically active. Interestingly, we demonstrate that three of these active M1 paralogs have roles independent of PAM-1 in promoting gametogenesis and fecundity. Simultaneous inhibition of pam-1 and M1 paralogs produces synergistic decreases in overall brood sizes and embryonic viability, exacerbates the germinal phenotypes of pachytene extension and delayed nucleolar disassembly, and unmasks previously hidden phenotypes. Our data suggests that the interdependent functions of multiple M1 aminopeptidases are necessary for reproductive success in C. elegans and lend further credence to the redundant composition of an evolutionarily conserved enzyme family.




Similar content being viewed by others
References
Abe T, Yamamoto Y, Hazato T (1998) Changes in aminopeptidase N located on neutrophils derived from patients with chronic pain. Masui 47(2):151–155
Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77(1):71–94
Brooks DR, Hooper NM, Isaac RE (2003) The Caenorhabditis elegans orthologue of mammalian puromycin-sensitive aminopeptidase has roles in embryogenesis and reproduction. J Biol Chem 278(44):42795–42801
Church DL, Guan KL, Lambie EJ (1995) Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development 121(8):2525–2535
Constam DB, Tobler AR, Rensing-Ehl A, Kemler I, Hersh LB, Fontana A (1995) Puromycin-sensitive aminopeptidase. Sequence analysis, expression, and functional characterization. J Biol Chem 270(45):26931–26939
Cooke J, Nowak MA, Boerlijst M, Maynard-Smith J (1997) Evolutionary origins and maintenance of redundant gene expression during metazoan development. Trends Genet 13(9):360–364
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36(Web Server issue):W465–W469
Fortin SM, Marshall SL, Jaeger EC, Greene PE, Brady LK, Isaac RE, Schrandt JC, Brooks DR, Lyczak R (2010) The PAM-1 aminopeptidase regulates centrosome positioning to ensure anterior-posterior axis specification in one-cell C. elegans embryos. Dev Biol 344(2):992–1000
Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408(6810):325–330
Hersh LB, McKelvy JF (1981) An aminopeptidase from bovine brain which catalyzes the hydrolysis of enkephalin. J Neurochem 36(1):171–178
Hui KS, Wang YJ, Lajtha A (1983) Purification and characterization of an enkephalin aminopeptidase from rat brain membranes. Biochemistry 22(5):1062–1067
Hui M, Budai ED, Lajtha A, Palkovits M, Hui KS (1995) Changes in puromycin-sensitive aminopeptidases in postmortem schizophrenic brain regions. Neurochem Int 27(4–5):433–441
Iturrioz X, Rozenfeld R, Michaud A, Corvol P, Llorens-Cortes C (2001) Study of asparagine 353 in aminopeptidase A: characterization of a novel motif (GXMEN) implicated in exopeptidase specificity of monozinc aminopeptidases. Biochemistry 40(48):14440–14448
Johnson GD, Hersh LB (1990) Studies on the subsite specificity of the rat brain puromycin-sensitive aminopeptidase. Arch Biochem Biophys 276(2):305–309
Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421(6920):231–237
Kudo LC, Parfenova L, Ren G, Vi N, Hui M, Ma Z, Lau K, Gray M, Bardag-Gorce F, Wiedau-Pazos M, Hui KS, Karsten SL (2011) Puromycin-sensitive aminopeptidase (PSA/NPEPPS) impedes development of neuropathology in hPSA/TAU(P301L) double-transgenic mice. Hum Mol Genet 20(9):1820–1833
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Laustsen PG, Vang S, Kristensen T (2001) Mutational analysis of the active site of human insulin-regulated aminopeptidase. Eur J Biochem 268(1):98–104
Lo MC, Gay F, Odom R, Shi Y, Lin R (2004) Phosphorylation by the beta-catenin/MAPK complex promotes 14-3-3-mediated nuclear export of TCF/POP-1 in signal-responsive cells in C. elegans. Cell 117(1):95–106
Lyczak R, Zweier L, Group T, Murrow MA, Snyder C, Kulovitz L, Beatty A, Smith K, Bowerman B (2006) The puromycin-sensitive aminopeptidase PAM-1 is required for meiotic exit and anteroposterior polarity in the one-cell Caenorhabditis elegans embryo. Development 133(21):4281–4292
Menzies FM, Hourez R, Imarisio S, Raspe M, Sadiq O, Chandraratna D, O'Kane C, Rock KL, Reits E, Goldberg AL, Rubinsztein DC (2010) Puromycin-sensitive aminopeptidase protects against aggregation-prone proteins via autophagy. Hum Mol Genet 19(23):4573–4586
Nowak MA, Boerlijst MC, Cooke J, Smith JM (1997) Evolution of genetic redundancy. Nature 388(6638):167–171
Osada T, Sakaki Y, Takeuchi T (1999) Puromycin-sensitive aminopeptidase gene (Psa) maps to mouse chromosome 11. Genomics 56(3):361–362
Osada T, Watanabe G, Kondo S, Toyoda M, Sakaki Y, Takeuchi T (2001a) Male reproductive defects caused by puromycin-sensitive aminopeptidase deficiency in mice. Mol Endocrinol 15(6):960–971
Osada T, Watanabe G, Sakaki Y, Takeuchi T (2001b) Puromycin-sensitive aminopeptidase is essential for the maternal recognition of pregnancy in mice. Mol Endocrinol 15(6):882–893
Pang KM, Ishidate T, Nakamura K, Shirayama M, Trzepacz C, Schubert CM, Priess JR, Mello CC (2004) The minibrain kinase homolog, mbk-2, is required for spindle positioning and asymmetric cell division in early C. elegans embryos. Dev Biol 265(1):127–139
Peer WA (2011) The role of multifunctional M1 metallopeptidases in cell cycle progression. Ann Bot 107(7):1171–1181
Pradillo M, Lopez E, Romero C, Sanchez-Moran E, Cunado N, Santos JL (2007) An analysis of univalent segregation in meiotic mutants of Arabidopsis thaliana: a possible role for synaptonemal complex. Genetics 175(2):505–511
Rawlings ND, Barrett AJ, Bateman A (2012) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 40(Database issue):D343–D350
Rutkowski R, Dickinson R, Stewart G, Craig A, Schimpl M, Keyse SM, Gartner A (2011) Regulation of Caenorhabditis elegans p53/CEP-1-dependent germ cell apoptosis by Ras/MAPK signaling. PLoS Genet 7(8):e1002238
Safavi A, Hersh LB (1995) Degradation of dynorphin-related peptides by the puromycin-sensitive aminopeptidase and aminopeptidase M. J Neurochem 65(1):389–395
Sanchez-Moran E, Jones GH, Franklin FC, Santos JL (2004) A puromycin-sensitive aminopeptidase is essential for meiosis in Arabidopsis thaliana. Plant Cell 16(11):2895–2909
Schomburg L, Turwitt S, Prescher G, Lohmann D, Horsthemke B, Bauer K (1999) Human TRH-degrading ectoenzyme cDNA cloning, functional expression, genomic structure and chromosomal assignment. Eur J Biochem FEBS 265(1):415–422
Schulz C, Perezgasga L, Fuller MT (2001) Genetic analysis of dPsa, the Drosophila orthologue of puromycin-sensitive aminopeptidase, suggests redundancy of aminopeptidases. Dev Genes Evol 211(12):581–588
Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, Hewitson M, Holz C, Khan M, Lazik S, Martin C, Nitzsche B, Ruer M, Stamford J, Winzi M, Heinkel R, Roder M, Finell J, Hantsch H, Jones SJ, Jones M, Piano F, Gunsalus KC, Oegema K, Gonczy P, Coulson A, Hyman AA, Echeverri CJ (2005) Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434(7032):462–469
Taylor A (1993) Aminopeptidases: structure and function. FASEB J 7:290–298
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25(24):4876–4882
Timmons L, Court DL, Fire A (2001) Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263(1–2):103–112
Timmons L, Fire A (1998) Specific interference by ingested dsRNA. Nature 395(6705):854
Tobler AR, Constam DB, Schmitt-Graff A, Malipiero U, Schlapbach R, Fontana A (1997) Cloning of the human puromycin-sensitive aminopeptidase and evidence for expression in neurons. J Neurochem 68(3):889–897
Towne CF, York IA, Neijssen J, Karow ML, Murphy AJ, Valenzuela DM, Yancopoulos GD, Neefjes JJ, Rock KL (2008) Puromycin-sensitive aminopeptidase limits MHC class I presentation in dendritic cells but does not affect CD8 T cell responses during viral infections. J Immunol 180(3):1704–1712
Tucker M, Han M (2008) Muscle cell migrations of C. elegans are mediated by the alpha-integrin INA-1, Eph receptor VAB-1, and a novel peptidase homologue MNP-1. Dev Biol 318(2):215–223
Vavouri T, Semple JI, Lehner B (2008) Widespread conservation of genetic redundancy during a billion years of eukaryotic evolution. Trends Genet 24(10):485–488
Walker AK, Shi Y, Blackwell TK (2004) An extensive requirement for transcription factor IID-specific TAF-1 in Caenorhabditis elegans embryonic transcription. J Biol Chem 279(15):15339–15347
Yamamoto Y, Li YH, Ushiyama I, Nishimura A, Ohkubo I, Nishi K (2000) Puromycin-sensitive alanyl aminopeptidase from human liver cytosol: purification and characterization. Forensic Sci Int 113(1–3):143–146
Yao T, Cohen RE (1999) Giant proteases: beyond the proteasome. Curr Biol CB 9(15):R551–R553
Ye S, Chai SY, Lew RA, Albiston AL (2007) Insulin-regulated aminopeptidase: analysis of peptide substrate and inhibitor binding to the catalytic domain. Biol Chem 388(4):399–403
Acknowledgments
We thank L. Samples, C. Wells, and A. Cude for the technical assistance and R. Lyczak for sharing the pam-1(or282) strain. This work was supported by the Kentucky EPSCoR (C.T.) and institutional funding from Murray State University (C.T.) and the Murray State University Office of Undergraduate Research and Scholarly Activities (M.J.A.).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: David A. Weisblat
Rights and permissions
About this article
Cite this article
Althoff, M.J., Flick, K. & Trzepacz, C. Collaboration within the M1 aminopeptidase family promotes reproductive success in Caenorhabditis elegans . Dev Genes Evol 224, 137–146 (2014). https://doi.org/10.1007/s00427-014-0470-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-014-0470-3