Abstract
Main conclusion
Genetically diverse cottonseeds show altered compositions and spatial distributions of phosphatidylcholines and triacylglycerols. Lipidomics profiling led to the discovery of a novel FAD2 - 1 allele, fad2 - 1D - 1 , resulting in a high oleic phenotype.
The domestication and breeding of cotton for elite, high‐fiber cultivars have led to reduced variation of seed constituents within currently cultivated upland cotton genotypes. However, a recent screen of the genetically diverse U.S. National Cotton Germplasm Collection identified Gossypium accessions with marked differences in seed oil and protein content. Here, several of these accessions representing substantial variation in seed oil content were analyzed for quantitative and spatial differences in lipid compositions by mass spectrometric approaches. Results indicate considerable variation in amount and spatial distribution of pathway metabolites for triacylglycerol biosynthesis in embryos across Gossypium accessions, suggesting that this variation might be exploited by breeders for seed composition traits. By way of example, these lipid metabolite differences led to the identification of a mutant allele of the D-subgenome homolog of the delta-12 desaturase (fad2-1D-1) in a wild accession of G. barbadense that has a high oil and high oleic seed phenotype. This mutation is a 90-bp insertion in the 3′ end of the FAD2-1D coding sequence and a modification of the 3′ end of the gene beyond the coding sequence leading to the introduction of a premature stop codon. Given the large amounts of cottonseed produced around the world that is currently not processed into higher value products, these efforts might be one avenue to raise the overall value of the cotton crop for producers.










Similar content being viewed by others
Abbreviations
- MALDI:
-
Matrix assisted laser desorption/ionization
- MS:
-
Mass spectrometry
- ESI:
-
Electron spray ionization
- TD‐NMR:
-
Time-domain 1H nuclear magnetic resonance
- PC:
-
Phosphatidylcholine
- TAG:
-
Triacylglycerol
- TLE:
-
Total lipid extract
- P:
-
Palmitic acid (16:0)
- O:
-
Oleic acid (18:1)
- L:
-
Linoleic acid (18:2)
- Numerical designation of lipids indicates number of carbons in acyl chains:
-
Number of double bonds
References
AOCS (2009a) Generic combustion method for determination of crude protein (Ba 4e-93). Official and recommended practices of the AOCS, 6 edn. AOCS Press, Champaign, IL
AOCS (2009b) Nitrogen–ammonia-protein modified kjeldahl method (Ba 4d-90). Official and recommended practices of the AOCS, 6 edn. AOCS Press, Champaign, IL
AOCS (2009c) Oil (Aa 4-38). Official and recommended practices of the AOCS, 6 edn. AOCS Press, Champaign, IL
Auld DL, Heikkinen MK, Erickson DA, Sernyk JL, Romero JE (1992) Rapeseed mutants with reduced levels of polyunsaturated fatty acids and increased levels of oleic acid. Crop Sci 32(3):657–662. doi:10.2135/cropsci1992.0011183X003200030016x
Bland JM, Conkerton EJ, Abraham G (1991) Triacylglyceride composition of cottonseed oil by HPLC and GC. J Am Oil Chem Soc 68(11):840–843
Chapman KD, Ohlrogge JB (2012) Compartmentation of triacylglycerol accumulation in plants. J Biol Chem 287(4):2288–2294. doi:10.1074/jbc.R111.290072
Dowd MK, Boykin DL, Meredith WR Jr, Campbell BT, Bourland FM, Gannaway JR, Glass KM, Zhang J (2010) Fatty acid profiles of cottonseed genotypes from the national cotton variety trials. J Cotton Sci 14:64–73
Dyer JM, Stymne S, Green AG, Carlsson AS (2008) High-value oils from plants. Plant J 54(4):640–655. doi:10.1111/j.1365-313X.2008.03430.x
Hinze LL, Horn PJ, Kothari N, Dever JK, Frelichowski J, Chapman KD, Percy RG (2015) Nondestructive measurements of cottonseed nutritional trait diversity in the US National cotton germplasm collection. Crop Sci 55(2):770–782. doi:10.2135/cropsci2014.04.0318
Horn PJ, Chapman KD (2012) Lipidomics in tissues, cells and subcellular compartments. Plant J 70(1):69–80. doi:10.1111/j.1365-313X.2011.04868.x
Horn PJ, Chapman KD (2014a) Lipidomics in situ: insights into plant lipid metabolism from high resolution spatial maps of metabolites. Prog Lipid Res 54:32–52. doi:10.1016/j.plipres.2014.01.003
Horn PJ, Chapman KD (2014b) Metabolite Imager: customized spatial analysis of metabolite distributions in mass spectrometry imaging. Metabolomics 10(2):337–348. doi:10.1007/s11306-013-0575-0
Horn PJ, Ledbetter NR, James CN, Hoffman WD, Case CR, Verbeck GF, Chapman KD (2011a) Visualization of lipid droplet composition by direct organelle mass spectrometry. J Biol Chem 286(5):3298–3306. doi:10.1074/jbc.M110.186353
Horn PJ, Neogi P, Tombokan X, Ghosh S, Campbell BT, Chapman KD (2011b) Simultaneous quantification of oil and protein in cottonseed by low-field time-domain nuclear magnetic resonance. J Am Oil Chem Soc 88(10):1521–1529. doi:10.1007/s11746-011-1829-5
Horn PJ, Korte AR, Neogi PB, Love E, Fuchs J, Strupat K, Borisjuk L, Shulaev V, Lee Y-J, Chapman KD (2012) Spatial mapping of lipids at cellular resolution in embryos of cotton. Plant Cell Online
Horn PJ, Sturtevant D, Chapman KD (2014) Modified oleic cottonseeds show altered content, composition and tissue-specific distribution of triacylglycerol molecular species. Biochimie 96:28–36. doi:10.1016/j.biochi.2013.08.010
Jiao X, Zhao X, Zhou XR, Green AG, Fan Y, Wang L, Singh SP, Liu Q (2013) Comparative transcriptomic analysis of developing cotton cotyledons and embryo axis. PLoS One 8(8):e71756. doi:10.1371/journal.pone.0071756
Jones LA, King CC (1996) Cottonseed oil. In: Bailey’s industrial oil and fat products, vol 2. Edible oil and fat products: oil and oil seeds, 5 edn. Wiley, New York
Kargiotidou A, Deli D, Galanopoulou D, Tsaftaris A, Farmaki T (2008) Low temperature and light regulate delta 12 fatty acid desaturases (FAD2) at a transcriptional level in cotton (Gossypium hirsutum). J Exp Bot 59(8):2043–2056. doi:10.1093/jxb/ern065
Kinney AJ (1996) Development of genetically engineered soybean oils for food applications. J Food Lipids 3(4):273–292. doi:10.1111/j.1745-4522.1996.tb00074.x
Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, DeBono A, Durrett TP, Franke RB, Graham IA, Katayama K, Kelly AA, Larson T, Markham JE, Miquel M, Molina I, Nishida I, Rowland O, Samuels L, Schmid KM, Wada H, Welti R, Xu C, Zallot R, Ohlrogge J (2013) Acyl-lipid metabolism. The Arabidopsis Book e0161. doi:10.1199/tab.0161
Lísa M, Holcapek M (2008) Triacylglycerols profiling in plant oils important in food industry, dietetics and cosmetics using high-performance liquid chromatography-atmospheric pressure chemical ionization mass spectrometry. J Chromatogr A 1198–1199:115–130. doi:10.1016/j.chroma.2008.05.037
Liu Q, Singh S, Green A (2000) Genetic modification of cotton seed oil using inverted-repeat gene-silencing techniques. Biochem Soc T 28:927–929. doi:10.1042/0300-5127:0280927
Liu Q, Singh S, Chapman K, Green A (2009) Bridging traditional and molecular genetics in modifying cottonseed oil. In: Paterson AH (ed) Genetics and genomics of cotton. Springer US, New York, NY, pp 353–382. doi:10.1007/978-0-387-70810-2_15
Liu Q, Llewellyn DJ, Singh SP, Green AG (2012) Cotton seed development: opportunities to add value to a byproduct of fiber production. Flower Fruit 133
McCartney AW, Dyer JM, Dhanoa PK, Kim PK, Andrews DW, McNew JA, Mullen RT (2004) Membrane-bound fatty acid desaturases are inserted co-translationally into the ER and contain different ER retrieval motifs at their carboxy termini. Plant J 37(2):156–173
Metcalfe LD, Schmitz AA, Pelka JR (1966) Rapid preparation of fatty acid esters from lipids for gas chromatographic analysis. Anal Chem 38(3):514–515
Miller JF, Zimmerman DC, Vick BA (1987) Genetic control of high oleic acid content in sunflower oil1. Crop Sci 27(5):923–926. doi:10.2135/cropsci1987.0011183X002700050019x
Norden AJ, Gorbet DW, Knauft DA, Young CT (1987) Variability in oil quality among peanut genotypes in the florida breeding program. Peanut Science 14(1):7–11. doi:10.3146/i0095-3679-14-1-3
Ohlrogge J, Browse J (1995) Lipid Biosynthesis. Plant Cell 7(7):957–970. doi:10.1105/tpc.7.7.957
Page JT, Huynh MD, Liechty ZS, Grupp K, Stelly D, Hulse AM, Ashrafi H, Van Deynze A, Wendel JF, Udall JA (2013) Insights into the evolution of cotton diploids and polyploids from whole-genome re-sequencing. G3-Genes Genom Genet 3(10):1809–1818. doi:10.1534/g3.113.007229
Pham AT, Lee JD, Shannon JG, Bilyeu KD (2011) A novel FAD2-1 A allele in a soybean plant introduction offers an alternate means to produce soybean seed oil with 85% oleic acid content. Theor Appl Genet 123(5):793–802. doi:10.1007/s00122-011-1627-3
Rathore KS, Sundaram S, Sunilkumar G, Campbell LM, Puckhaber L, Marcel S, Palle SR, Stipanovic RD, Wedegaertner TC (2012) Ultra-low gossypol cottonseed: generational stability of the seed-specific, RNAi-mediated phenotype and resumption of terpenoid profile following seed germination. Plant Biotechnol J 10(2):174–183. doi:10.1111/j.1467-7652.2011.00652.x
Shanklin J, Cahoon EB (1998) Desaturation and related modifications of fatty acids. Ann Rev Plant Physiol Plant Mol Biol 49(1):611–641. doi:10.1146/annurev.arplant.49.1.611
Shockey J, Dowd M, Mack B, Gilbert M, Scheffler B, Ballard L, Frelichowski J, Mason C (2016) Naturally occurring high oleic acid cottonseed oil: identification and functional analysis of a mutant allele of Gossypium barbadense fatty acid desaturase-2. Planta. doi:10.1007/s00425-016-2633-0
Sturtevant D, Lee YJ, Chapman KD (2016) Matrix assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI) for direct visualization of plant metabolites in situ. Curr Opin Biotechnol 37:53–60. doi:10.1016/j.copbio.2015.10.004
Sunilkumar G, Campbell LM, Puckhaber L, Stipanovic RD, Rathore KS (2006) Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc Natl Acad Sci USA 103(48):18054–18059. doi:10.1073/pnas.0605389103
Tiwari GJ, Liu Q, Shreshtha P, Li Z, Rahman S (2016) RNAi-mediated down-regulation of the expression of OsFAD2-1: effect on lipid accumulation and expression of lipid biosynthetic genes in the rice grain. BMC Plant Biol 16(1):189. doi:10.1186/s12870-016-0881-6
Yunusova SG, Gusakova SD, Glushenkova AI, Nadzhimov UK, Turabekov S, Musaev SA (1991) A comparative investigation of the fatty acid compositions of the seeds of a number of lines of a genetic collection of Gossypium hirsutum. Chem Nat Compd 27(2):147–150
Zhang D, Pirtle IL, Park SJ, Nampaisansuk M, Neogi P, Wanjie SW, Pirtle RM, Chapman KD (2009) Identification and expression of a new delta-12 fatty acid desaturase (FAD2-4) gene in upland cotton and its functional expression in yeast and Arabidopsis thaliana plants. Plant Physiol Biochem 47(6):462–471. doi:10.1016/j.plaphy.2008.12.024
Acknowledgements
This research was supported in part by grants from Cotton Incorporated (Agreement# 08‐395) to screen cotton germplasm. We thank the Hoblitzelle Foundation for the support of MS imaging and cryostat instrumentation. We thank Drs. Vladimir Shulaev and Guido Verbeck, University of North Texas, for ongoing analytical advice. We also thank Drs. Kerstin Strupat and Mari Prieto Conaway of Thermo‐Fisher Scientific for technical support in MS imaging experiments. Patrick Horn was supported through the UNT Doctoral Fellowship program.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2016_2630_MOESM2_ESM.png
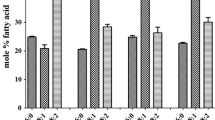
Supplemental Fig. 1. Fatty acid composition from extracts of Gossypium accessions. Relative quantification of fatty acids on a mol % were measuring palmitic (16:0; P), steric (18:0; S), oleic (18:1: O), and linoleic (18:2; L) fatty acids. Measurements were taken in triplicate with 5 seeds each in each replicate (PNG 107 kb)
425_2016_2630_MOESM3_ESM.png
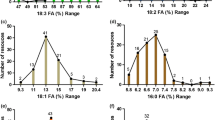
Supplemental Fig. 2. Images of lipid metabolites in cross-sections of embryos (middle) from G. barbadense (GB-0331). Ion maps are generated from MALDI-MS for selected PC (a) and TAG (b) molecular species containing one cyclic fatty acid (either sterculic (19:1) or dihydrosterculic (19:0); malvalic acid, also a cyclic fatty acid in cottonseed cannot be resolved from oleic acid in these analyses and so species with malvalic acid are not shown). The intensity scale in all rows are set to the same values so direct comparison can be made for all PC and TAG molecular species respectively. PC and TAG molecular species are denoted as total number of acyl carbons and number of total double bonds. Images are converted from mol % of class with red as highest relative amount. For orientation, a brightfield image of the longitudinal section prior to matrix application and MS imaging is provided in (c) (PNG 2552 kb)
425_2016_2630_MOESM4_ESM.png
Supplemental Fig. 3. PCR amplification of FAD2-1A and FAD2-1D in G. barbadense accessions. (a) Agarose gel electrophoresis of genomic DNA fragments with gene specific primers of the A-homolog of FAD2-1 in G. barbadense backgrounds. (b) Agarose gel electrophoresis of genomic DNA fragments with gene specific primers for long-read PCR spanning FAD2-1 (5′) + Gorai.013G248700 (3′) in G. barbadense backgrounds.. The long-read PCR product in GB-0331 was ~ 10 Kb, while in Pima-S6 it was ~ 7 Kb. Gene specific primers used for amplification can be found in Supplementary Table 1. (c) Agarose gel electrophoresis of RT-PCR products of FAD2-1D and Actin control common coding sequence gene specific primers in G. barbadense backgrounds. Primers for FAD2-1 were “FAD2-1D common coding seq” (Table S1). Pima-S7 was used here as an additional negative control against GB-0331. (d) Schematic of the long-read primer design spanning FAD2-1D and Gorai.013G248700-D (PNG 746 kb)
Rights and permissions
About this article
Cite this article
Sturtevant, D., Horn, P., Kennedy, C. et al. Lipid metabolites in seeds of diverse Gossypium accessions: molecular identification of a high oleic mutant allele. Planta 245, 595–610 (2017). https://doi.org/10.1007/s00425-016-2630-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2630-3