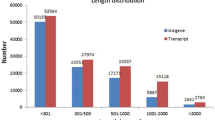
Abstract
Main conclusion
New molecular markers were developed for targeting Thinopyrum intermedium 1St#2 chromosome, and novel FISH probe representing the terminal repeats was produced for identification of Thinopyrum chromosomes.
Thinopyrum intermedium has been used as a valuable resource for improving the disease resistance and yield potential of wheat. A wheat-Th. intermedium ssp. trichophorum chromosome 1St#2 substitution and translocation has displayed superior grain protein and wet gluten content. With the aim to develop a number of chromosome 1St#2 specific molecular and cytogenetic markers, a high throughput, low-cost specific-locus amplified fragment sequencing (SLAF-seq) technology was used to compare the sequences between a wheat-Thinopyrum 1St#2 (1D) substitution and the related species Pseudoroegneria spicata (St genome, 2n = 14). A total of 5142 polymorphic fragments were analyzed and 359 different SLAF markers for 1St#2 were predicted. Thirty-seven specific molecular markers were validated by PCR from 50 randomly selected SLAFs. Meanwhile, the distribution of transposable elements (TEs) at the family level between wheat and St genomes was compared using the SLAFs. A new oligo-nucleotide probe named Oligo-pSt122 from high SLAF reads was produced for fluorescence in situ hybridization (FISH), and was observed to hybridize to the terminal region of 1St#L and also onto the terminal heterochromatic region of Th. intermedium genomes. The genome-wide markers and repetitive based probe Oligo-pSt122 will be valuable for identifying Thinopyrum chromosome segments in wheat backgrounds.





Similar content being viewed by others
References
AACC (2000) Approved methods of the American Association of Cereal Chemists, 10th edn. American Association of Cereal Chemists, St Paul
Anugrahwati DR, Shepherd KW, Verlin DC, Zhang P, Mirzaghaderi G, Walker E, Francki MG, Dundas IS (2008) Isolation of wheat-1RS recombinants which break the linkage between stem rust resistance gene SrR and secalin. Genome 51:341–349
Bartos B (1993) Chromosome 1R of rye in wheat breeding. Plant Breed Abstr 63:1203–1211
Biscotti MA, Canapa A, Forconi M, Olmo E, Barucca M (2015) Transcription of tandemly repetitive DNA: functional roles. Chromosome Res 23:463–477
Brandes A, Roder MS, Ganal MW (1995) Barley telomeres are associated with two different types of satellite DNA sequences. Chromosome Res 3:315–320
Cai X, Jones SS, Murray TD (1998) Molecular cytogenetic characterization of Thinopyrum and wheat-Thinopyrum translocated chromosomes in a wheat-Thinopyrum amphiploid. Chromosome Res 6:183–189
Chen Q (2005) Detection of alien chromatin introgression from Thinopyrum into wheat using S genomic DNA as a probe—A landmark approach for Thinopyrum genome research. Cytogenet Genome Res 109:350–359
Chen Q, Conner RL, Laroche A, Thomas JB (1998) Genome analysis of Thinopyrum intermedium and Th. ponticum using genomic in situ hybridization. Genome 141:580–586
Chen S, Huang Z, Dai Y, Qin S, Gao Y, Zhang L, Gao Y, Chen J (2013) The development of 7E chromosome-specific molecular markers for Thinopyrum elongatum based on SLAF-seq technology. PLoS One 8:e65122
Cheng ZK, Stupar RM, Gu MH, Jiang JM (2001) A tandemly repeated DNA sequence is associated with both knob-like heterochromatin and a highly decondensed structure in the meiotic pachytene chromosomes of rice. Chromosoma 110:24–31
Cuadrado A, Rubio P, Ferrer E, Jouve N (1996) Sequential combinations of C-banding and in situ hybridization and their use in the detection of interspecific introgressions into wheat. Euphytica 89:107–112
De Pace C, Snidaro D, Ciaffi M, Vittori D, Ciofo A, Cenci A, Tanzarella OA, Qualset CO, Scarascia Mugnozza GT (2001) Introgression of Dasypyrum villosum chromatin into common wheat improves grain protein quality. Euphytica 117:67–75
Dechyeva D, Schmidt T (2006) Molecular organization of terminal repetitive DNA in Beta species. Chromosome Res 14:881–897
Edwards D, Batley J (2010) Plant genome sequencing: applications for crop improvement. Plant Biotechnol J 7:1–8
Edwards D, Batley J, Snowdon RJ (2013) Accessing complex crop genomes with next-generation sequencing. Theor Appl Genet 126:1–11
Friebe B, Mukai Y, Gill BS, Cauderon Y (1992) C-banding and in situ hybridization analyses of Agropyron intermedium, a partial wheat-Ag. intermedium amphiploid, and six derived chromosome addition lines. Theor Appl Genet 84:899–905
Fu S, Chen L, Wang Y, Li M, Yang Z, Qiu L, Yan B, Ren Z, Tang Z (2015) Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci Rep 5:10552
Garg M, Tanaka H, Ishikawa N, Takata K, Yanaka M, Tsujimoto H (2009a) Agropyron elongatum HMW-glutenins have the potential to improve wheat end product quality through targeted chromosome introgression. J Cereal Sci 50(3):358–363
Garg M, Tanaka H, Tsujimoto H (2009b) Exploration of Triticeae seed storage proteins for improvement of wheat end product quality. Breed Sci 59:519–528
Han FP, Lamb JC, Birchler JA (2006) High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Pro Natl Acad Sci USA 103:3238–3243
Han H, Bai L, Su J, Zhang J, Song L, Gao A, Yang X, Li X, Liu W, Li L (2014) Genetic rearrangements of six wheat-Agropyron cristatum 6P addition lines revealed by molecular markers. PLoS One 9:e91066
Heslop-Harrison JS (2000) Comparative genome organization in plants: from sequence and markers to chromatin and chromosomes. Plant Cell 12:617–636
Heslop-Harrison JS, Schwarzacher T (2011) Organisation of the plant genome in chromosomes. Plant J 66:18–33
Howell T, Hale I, Jankuloski L, Bonafede M, Gilbert M, Dubcovsky J (2014) Mapping a region within the 1RS.1BL translocation in common wheat affecting grain yield and canopy water status. Theor Appl Genet 127:2695–2709
Hu LJ, Li GR, Zeng ZX, Chang ZJ, Liu C, Zhou JP, Yang ZJ (2011) Molecular cytogenetic identification of a new wheat-Thinopyrum substitution line with stripe rust resistance. Euphytica 177:169–177
Hu LJ, Li GR, Zhan HX, Liu C, Yang ZJ (2012) New St-chromosome specific molecular markers for identifying wheat-Thinopyrum intermedium derivative lines. J Genet 91:e69–e74
Hurni S, Brunner S, Stirnweis D, Herren G, Peditto D, McIntosh RA, Keller B (2014) The powdery mildew resistance gene Pm8 derived from rye is suppressed by its wheat ortholog Pm3. Plant J 79:904–913
Jauhar PP, Peterson TS, Xu SS (2009) Cytogenetic and molecular characterization of a durum alien disomic addition line with enhanced tolerance to Fusarium head blight. Genome 52:467–483
Jiang J, Gill BS (1993) Sequential chromosome banding and in situ hybridization analysis. Genome 36:792–795
Kishii M, Nagaki K, Tsujimoto H, Sasakuma T (1999) Exclusive localization of tandem repetitive sequences in subtelomeric heterochromatin regions of Leymus racemosus (Poaceae, Triticeae). Chromosome Res 7:519–529
Li H, Wang X (2009) Thinopyrum ponticum and the promising source of resistance to fungal and viral diseases of wheat. J Genet Genomics 36:557–565
Li WL, Chen PD, Qi LL, Liu DJ (1995) Isolation, characterization and application of a species-specific repeated sequence from Haynaldia villosa. Theor Appl Genet 90:526–533
Li WL, Zhang P, Fellers JP, Friebe B, Gill BS (2004) Sequence composition, organization, and evolution of the core Triticeae genome. Plant J 40:500–511
Li Y, Song Y, Zhou R, Branlard G, Jia J (2009) Detection of QTLs for bread-making quality in wheat using a recombinant inbred line population. Plant Breed 128:235–243
Li GR, Liu C, Li CH, Zhao JM, Zhou L, Dai G, Yang EN, Yang ZJ (2013) Introgression of a novel Thinopyrum intermedium St-chromosome-specific HMW-GS gene into wheat. Mol Breed 31:843–853
Li G, Lang T, Dai G, Li D, Li C, Song X, Yang Z (2015a) Precise identification of two wheat-Thinopyrum intermedium substitutions reveals the compensation and rearrangement between wheat and Thinopyrum chromosomes. Mol Breed 35:1
Li G, Zhang H, Zhou L, Gao D, Lei M, Zhang J, Yang Z (2015b) Molecular characterization of Sec2 loci in wheat-Secale africanum derivatives demonstrates genomic divergence of Secale species. Int J Mol Sci 16(4):8324–8336
Li G, Gao D, Zhang H, Li J, Wang H, La S, Ma J, Yang Z (2016) Molecular cytogenetic characterization of Dasypyrum breviaristatum chromosomes in wheat background revealing the genomic divergence between Dasypyrum species. Mol Cytogenet 9:6
Liu C, Yang ZJ, Jia JQ, Li GR, Zhou JP, Ren ZL (2009) Genomic distribution of a long terminal repeat (LTR) Sabrina-like retrotransposon in Triticeae species. Cereal Res Commun 37:363–372
Macas J, Neumann P, Navrátilová A (2007) Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics 8:427
Mago R, Zhang P, Vautrin S, Šimková H, Bansal U, Luo MC, Rouse M, Karaoglu H, Periyannan S, Kolmer J, Jin Y, Ayliffe MA, Bariana H, Park RF, McIntosh R, Doležel J, Bergès H, Spielmeyer B, Lagudah ES, Ellis JG, Dodds PN (2015) The wheat Sr50 gene reveals rich diversity at a cereal disease resistance locus. Nat Plant 1:15186
Mahelka V, Kopecky D, Pastova L (2011) On the genome constitution and evolution of intermediate wheatgrass (Thinopyrum intermedium: Poaceae, Triticeae). BMC Evol Biol 11:127
Mahelka V, Kopecky D, Baum BR (2013) Contrasting patterns of evolution of 45S and 5S rDNA families uncover new aspects in the genome constitution of the agronomically important grass Thinopyrum intermedium (Triticeae). Mol Biol Evol 30:2065–2086
Middleton C, Stein N, Keller B, Kilian B, Wicker T (2012) Comparative analysis of genome composition in Triticeae reveals strong variation in transposable element dynamics and nucleotide diversity. Plant J 73:347–356
Niu ZX, Klindworth DL, Wang RRC, Jauhar PP, Larkin PJ, Xu SS (2011) Characterization of HMW glutenin subunits in Thinopyrum intermedium, Th. bessarabicum, Lophopyrum elongatum, Aegilops markgrafii, and their addition lines in wheat. Crop Sci 51:667–677
Novak P, Neumann P, Macas J (2010) Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11:378
Peng JH, Zadeh H, Lazo GR, Gustafson JP, Chao S, Anderson OD, Qi LL, Echalier B, Gill BS, Dilbirligi M, Sandhu D, Gill KS, Greene RA, Sorrells ME, Akhunov ED, Dvorák J, Linkiewicz AM, Dubcovsky J, Hossain KG, Kalavacharla V, Kianian SF, Mahmoud AA, Miftahudin Conley EJ, Anderson JA, Pathan MS, Nguyen HT, McGuire PE, Qualset CO, Lapitan NL (2004) Chromosome bin map of expressed sequence tags in homoeologous group 1 of hexaploid wheat and homoeology with rice and Arabidopsis. Genetics 168:609–623
Pretorius ZA, Singh RP, Wagoire WW, Payne TS (2000) Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f. sp. tritici in Uganda. Plant Dis 84:203
Rabinovich SV (1998) Importance of wheat-rye translocations for breeding modern cultivars of Triticum aestivum L. Euphytica 100:323–340
Senerchia N, Wicker T, Felber F, Parisod C (2013) Evolutionary dynamics of retrotransposons assessed by high-throughput sequencing in wild relatives of wheat. Genome Biol Evol 5:1010–1020
Sergeeva EM, Salina EA, Adonina IG, Chalhoub B (2010) Evolutionary analysis of the CACTA DNA-transposon Caspar across wheat species using sequence comparison and in situ hybridization. Mol Genet Genomics 284:11–23
Stein N (2007) Triticeae genomics: advances in sequence analysis of large genome cereal crops. Chromosome Res 15:21–31
Sun X, Liu D, Zhang X, Li W, Liu H, Hong W, Jiang C, Guan N, Ma C, Zeng H (2013) SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS One 8:e58700
Tang ZX, Yang ZJ, Fu SL, Yang MY, Li GR, Zhang HQ, Tan FQ, Ren ZL (2011) A new long terminal repeat (LTR) sequence allows to identify J genome from JS and St genomes of Thinopyrum intermedium. J Appl Genet 52:31–33
Tang ZX, Yang ZJ, Fu SL (2014) Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J Appl Genet 55:313–318
Torres GA, Gong Z, Iovene M, Hirsch CD, Buell CR, Bryan GJ, Novák P, Macas J, Jiang J (2011) Organization and evolution of subtelomeric satellite repeats in the potato genome. G3 (Bethesda) 1:85–92
Van Deynze AE, Sorrells ME, Park WD, Ayres NM, Fu H, Cartinhour SW, Paul E, McCouch SR (1998) Anchor probes for comparative mapping in grass genera. Theor Appl Genet 97:356–369
Vershinin AV, Schwarzacher T, Heslop-Harrison HS (1995) The large-scale genomic organization of repetitive DNA families at the telomeres of rye chromosomes. Plant Cell 7:1823–1833
Wang Y, Wang H (2016) Molecular cytogenetic characterization of three novel wheat-Thinopyrum intermedium addition lines with novel storage protein subunits and resistance to both powdery mildew and stripe rust. J Genet Genomics 43:45–48
Wang RR, Zhang JY, Lee BS, Jensen KB, Kishii M, Tsujimoto H (2006) Variations in abundance of 2 repetitive sequences in Leymus and Psathyrostachys species. Genome 49:511–519
Wang RR, Larson SR, Jensen KB, Bushman BS, DeHaan LR, Wang S, Yan X (2015) Genome evolution of intermediate wheatgrass as revealed by EST-SSR markers developed from its three progenitor diploid species. Genome 58:63–70
Wicker T, Matthews DE, Keller B (2002) TREP: a database for Triticeae repetitive elements. Trends Plant Sci 7:561–562
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Wicker T, Taudien S, Houben A, Keller B, Graner A, Platzer M, Stein N (2009) A whole-genome snapshot of 454 sequences exposes the composition of the barley genome and provides evidence for parallel evolution of genome size in wheat and barley. Plant J 59:712–722
Xia C, Chen LL, Rong TZ, Li R, Xiang Y, Wang P, Liu CH, Dong XQ, Liu B, Zhao D, Wei RJ, Lan H (2015) Identification of a new maize inflorescence meristem mutant and association analysis using SLAF-seq method. Euphytica 202:35–44
Xu J, Conner RL (1994) Intravarietal variation in satellites and C-banded chromosomes of Agropyron intermedium ssp. trichophorum cv. Greenleaf. Genome 37:305–310
Yang ZJ, Li GR, Chang ZJ, Zhou JP, Ren ZL (2006) Characterization of a partial amphiploid between Triticum aestivum cv. Chinese Spring and Thinopyrum intermedium ssp. trichophorum. Euphytica 149:11–17
Yang ZJ, Li GR, Jia JQ, Zeng X, Lei MP, Zeng ZX, Zhang T, Ren ZL (2009) Molecular cytogenetic characterization of wheat- Secale africanum amphiploids and the introgression lines for stripe rust resistance. Euphytica 167(2):197–202
Zhang Y, Wang L, Xin H, Li D, Ma C, Ding X, Hong W, Zhang X (2013) Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Plant Biol 13:141
Zhang R, Zhang M, Wang X, Chen P (2014) Introduction of chromosome segment carrying the seed storage protein genes from chromosome 1V of Dasypyrum villosum showed positive effect on bread-making quality of common wheat. Theor Appl Genet 127:523–533
Zhang Y, Zhang J, Huang L, Gao A, Zhang J, Yang X, Liu W, Li X, Li L (2015) A high-density genetic map for P genome of Agropyron Gaertn. based on specific-locus amplified fragment sequencing (SLAF-seq). Planta 242:1335–1347
Acknowledgments
We particularly thank Dr. I. Dundas at the University of Adelaide, Australia for reviewing the manuscript, and Dr. Cheng Liu of Shandong Academy of Agricultural Sciences, China for seeds quality tests. We are thankful to the National Natural Science Foundation of China (No. 31171542 and 31101143), Applied and Basic Project (2016JY0075) from Science and Technology Department of Sichuan, China for their financial support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Li, G., Wang, H., Lang, T. et al. New molecular markers and cytogenetic probes enable chromosome identification of wheat-Thinopyrum intermedium introgression lines for improving protein and gluten contents. Planta 244, 865–876 (2016). https://doi.org/10.1007/s00425-016-2554-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2554-y