Abstract
Protein–protein interactions are at the core of cellular interactomics and are essential for various biological functions. Since proteins commonly function as macromolecular complexes, it is important to identify their interacting partners to better understand their function and the significance in these interactions. The acyl-CoA-binding proteins (ACBPs) of eukaryotes show conservation in the presence of a lipid-binding acyl-CoA-binding domain. In Arabidopsis thaliana, four of six members from the AtACBP family possess ankyrin repeats (AtACBP1 and AtACBP2) or kelch motifs (AtACBP4 and AtACBP5), which can potentially mediate protein–protein interactions. Through yeast two-hybrid screens, a dozen putative protein partners interacting with AtACBPs have been isolated from an Arabidopsis cDNA library. Investigations in the past decade on the interaction between AtACBPs and their protein partners have revealed novel roles for AtACBPs, including functions in mediating oxidative stress responses, heavy metal tolerance and oxygen sensing. Recent progress and current questions on AtACBPs and their interactors are discussed in this review.
Similar content being viewed by others
Introduction
Acyl-CoA-binding proteins (ACBPs) have been reported to bind long-chain acyl-CoA esters (C12-C22) through their acyl-CoA-binding domains with high specificities and affinities (Rasmussen et al. 1993; Chye 1998; Chye et al. 2000; Knudsen et al. 2000; Burton et al. 2005; Leung et al. 2004, 2006). The first identified ACBP was a neuropeptide (diazepam-binding inhibitor) from rat brain that inhibits benzodiazepine-binding (Guidotti et al. 1983). Subsequently, similar small cytosolic ACBPs were isolated from many eukaryotes and some prokaryotes (Burton et al. 2005; Xiao and Chye 2011a). These 10-kDa ACBPs were found to participate in acyl-CoA transport (Rasmussen et al. 1994; Schjerling et al. 1996) and in the maintenance of intracellular acyl-CoA pools (Knudsen et al. 1994; Gaigg et al. 2001; Huang et al. 2005), in membrane biosynthesis (Gaigg et al. 2001; Faergeman et al. 2004) and in the regulation of gene expression and enzyme activities related to lipid metabolism (Rasmussen et al. 1993; Mandrup et al. 1998; Kannan et al. 2003; Feddersen et al. 2007; Oikari et al. 2008).
In Arabidopsis thaliana, five larger ACBPs, AtACBP1 to AtACBP5, have been characterized besides the 10-kDa ACBP homologue, which has been designated as AtACBP6 (Xiao and Chye 2009, 2011a). Although Arabidopsis ACBPs are conserved at the acyl-CoA-binding domain, they show a range in size from 10.4 to 73.2 kDa (Fig. 1) (Xiao and Chye 2009, 2011a). AtACBP1 and its highly conserved homologue, AtACBP2, share 76.9 % amino acid identity and each has an N-terminal hydrophobic transmembrane domain and C-terminal ankyrin repeats (Fig. 1) (Chye 1998; Chye et al. 1999, 2000). AtACBP1 and AtACBP2 are expressed in siliques, stems, leaves, flowers and roots (Chye 1998; Chye et al. 2000). Previous studies have confirmed that they are subcellularly localized to the plasma membrane (PM) and the endoplasmic reticulum (ER) (Chye 1998; Chye et al. 1999, 2000). AtACBP3 has an N-terminal signal peptide and a transmembrane domain, and is targeted to the extracellular space (Fig. 1) (Leung et al. 2006). Kelch-motif containing AtACBP4 and AtACBP5 share 81.4 % amino acid identity (Fig. 1) (Leung et al. 2004). They are subcellularly localized to the cytosol (Xiao et al. 2008), while AtACBP4 has also been detected at the periphery of nuclei (Li et al. 2008). The 10-kDa AtACBP6, consisting solely of an ACB domain, was experimentally verified to be a cytosolic protein (Fig. 1) (Chen et al. 2008).
Domain architecture of Arabidopsis ACBPs. The numbers indicate amino acid positions in each protein. Six members of Arabidopsis ACBPs (AtACBPs) are distributed across four classes, classes I–IV, as indicated (Meng et al. 2011)
In recent years, increasing evidence has revealed that AtACBPs are involved in many biological processes, such as plant development (Chen et al. 2010; Du et al. 2013a), leaf senescence (Xiao et al. 2010), pathogen resistance (Xiao and Chye 2011b) and response to environmental stresses (Chen et al. 2008; Du et al. 2010, 2013b) including oxidative and heavy metal stresses (Xiao et al. 2008; Gao et al. 2009, 2010). In addition, AtACBPs containing ankyrin repeats (AtACBP1 and AtACBP2) and kelch motifs (AtACBP4) have been observed to interact with protein partners, some of which have also turned out to be stress-responsive proteins (Li and Chye 2004; Li et al. 2008; Gao et al. 2009, 2010; Licausi et al. 2011; Du et al. 2013a).
AtACBPs interact with enzymes of lipid metabolism
Ankyrin repeats are one of the most common motifs that mediate protein–protein interactions and are found in AtACBP1 and AtACBP2 (Fig. 1) (Michaely and Bennett 1992; Sedgwick and Smerdon 1999; Li et al. 2006). AtACBP2 has been reported to interact with LYSOPHOSPHOLIPASE2 (lysoPL2), which can detoxify lysophosphatidylcholine (lysoPC) (Gao et al. 2010). Lysophopholipids are generated by phospholipase A in response to stresses (Ryu 2004). Fluorescence resonance energy transfer (FRET) analysis using agroinfiltrated tobacco leaves revealed that the interaction between lysoPL2 and AtACBP2 occurs at the ER as well as the plasma membrane, via the ankyrin repeats of AtACBP2, consistent with the observations in yeast two-hybrid and co-immunoprecipitation assays (Gao et al. 2010). A mutant of AtACBP2 lacking the ankyrin repeat domain did not show interaction with lysoPL2 (Gao et al. 2010). The overexpression of lysoPL2 in transgenic Arabidopsis conferred enhanced tolerance to cadmium (Cd) and hydrogen peroxide (H2O2) treatments (Gao et al. 2010). Consistently, lysoPL2 mutants were sensitive to zinc (Zn) and H2O2, suggesting a positive role for lysoPL2 in oxidative stress (Gao et al. 2010). Subsequently, recombinant lysoPL2 was demonstrated to degrade toxic lysoPC in vitro (Gao et al. 2010). Considering that AtACBP2 binds lysoPC though its ACB domain and interacts with lysoPL2 via its ankyrin repeats, these partners likely promote lysoPC degradation (Fig. 2a).
Arabidopsis ACBPs interact with enzymes involved in lipid metablism. a AtACBP2 binds both lysoPL2 and lysoPC. AtACBP2 recruits lysoPL2 to degrade toxic lysoPC resulting from Zn or Cd-induced stress, b AtACBP1 binds PLDαl and PA. The membrane-associated PA likely inhibits ABI1 activity to promote ABA signaling. PA, phosphatidic acid; PC, phosphatidylcholine; PM, plasma membrane; lysoPC, lysophosphatidylcholine; ACB, acyl-CoA-binding domain; ANK, ankyrin repeats
Another ankyrin repeat-containing AtACBP, AtACBP1, has been recently reported to interact with PHOSPHOLIPASE Dα1 (PLDα1) at the plasma membrane by bimolecular fluorescence complementation (BiFC) and yeast two-hybrid assays (Du et al. 2013a). PLDα1 is a membrane phospholipase, producing phosphatidic acid (PA) (Fan et al. 1999), and its activity can be stimulated by abscisic acid (ABA) (Zhang et al. 2004). It has been shown that AtACBP1 overexpression in Arabidopsis enhances PLDα1 expression in response to ABA treatment (Du et al. 2013a). Given that recombinant AtACBP1 binds PA and phosphatidylcholine (PC) in vitro (Chen et al. 2010; Du et al. 2010, 2013a) and that AtACBP1 interacts with PLDα1 at the plasma membrane (Du et al. 2013a), AtACBP1 possibly promotes hydrolysis of PC to PA as a result of an increase in PLDα1 expression (Fig. 2b). Similar PC/PA changes occur in freezing stress; upregulation of AtACBP1 enhances PLDα1 expression in response to freezing stress and AtACBP1-overexpressors produce more PA but less PC than the wild type and the acbp1 mutant (Du et al. 2010). PLDα1-derived PA directly binds to ABSCISIC ACID-INSENSITIVE1 (ABI1), a principal negative regulator of ABA responses (Gosti et al. 1999; Merlot et al. 2001), leading to the suppression of ABI1 activity and thereby promotion of ABA signaling (Fig. 2b) (Zhang et al. 2004; Li et al. 2009). This model can explain the increase in ABA sensitivity of AtACBP1-overexpressors during seed germination and seedling development (Fig. 2b) (Du et al. 2013a).
AtACBPs interact with transcriptional factors
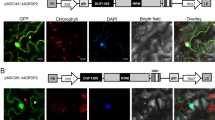
Besides enzymes in lipid metabolism, AtACBPs have been also noted to interact with transcriptional factors (TFs), including members from the ethylene response factor (ERF) family (Li and Chye 2004; Li et al. 2008; Licausi et al. 2011). Using yeast two-hybrid screening with an Arabidopsis cDNA library, AtACBP2 was first observed to interact with an ethylene-responsive element-binding protein, AtEBP, also named RELATED TO APETALA2.3 (RAP2.3) (Li and Chye 2004). Their interaction was further confirmed by co-immunoprecipitation assays and co-localization analysis by confocal microscopy, which revealed their interaction at the plasma membrane (Fig. 3) (Li and Chye 2004). This represented the first report suggesting that an AtACBP which is targeted to the ER and plasma membrane interacts with a TF (Li and Chye 2004).
Arabidopsis ACBPs interact with transcriptional factors. AtACBP2 and AtACBP4 may be involved in plant defense responses by their interactions with RAP2.3 at the plasma membrane and the cytosol, respectively (Li and Chye 2004; Li et al. 2008). AtACBP1 and AtACBP2 participate in oxygen sensing via interactions with RAP2.12 at the plasma membrane (Licausi et al. 2011). Under normoxia, RAP2.12 is sequestered by AtABCP1 or AtABCP2 at the plasma membrane (Licausi et al. 2011), and is thereby protected against degradation by the N-end rule pathway (Gibbs et al. 2011; Licausi et al. 2011). Upon hypoxia, RAP2.12 moves into the nucleus to activate the expression of hypoxia-responsive genes (Licausi et al. 2011). AtACBP1 also interacts with AREB1 via a yet unknown mechanism (Tse 2005)
Subsequently, another AtACBP, the cytosolic AtACBP4, was reported to interact with the same ERF, RAP2.3, in the cytosol (Fig. 3) (Li et al. 2008). This interaction, initially detected using yeast two-hybrid screens, was further confirmed by co-immunoprecipitation assays and FRET analysis (Li et al. 2008). It is interesting that both AtACBP2 and AtACBP4 interact with RAP2.3. Further investigations showed that it is the ankyrin repeats of AtACBP2 that facilitate the interaction between AtACBP2 and RAP2.3, because a deletion mutant of AtACBP2 lacking the ankyrin repeats failed to interact with RAP2.3 (Li and Chye 2004). In contrast, the interacting sites on AtACBP4 and RAP2.3 are still unclear (Li et al 2008). However, as shown in Fig. 1, AtACBP4 possesses kelch motifs which are known to mediate protein–protein interactions (Adams et al. 2000; Andrade et al. 2001). Thus, it has been predicted that AtACBP4 could interact with RAP2.3 via these motifs (Li et al. 2008). Given that the expression of RAP2.3 and AtACBP4 are induced by Botrytis cinerea infection, as well as by compounds that are essential in defense responses to B. cinerea, the ethylene precursor 1-aminocyclopropane-1-carboxylic acid and methyl jasmonate, RAP2.3 and AtACBP2/AtACBP4 may participate in plant defense responses through ethylene and/or jasmonate signaling (Fig. 3) (Li et al. 2008). Subsequently, interaction between RAP2.12 (which also belongs to the ERF family) and AtACBP1/AtACBP2 were reported using BiFC and yeast two-hybrid assays (Licausi et al. 2011).
The Arabidopsis ERF family includes over 100 members phylogenetically parsed into ten clades, while both RAP2.12 and RAP2.3 belong to the group VII ERFs (Nakano et al. 2006). There are five members in this group, RAP2.12, RAP2.2, RAP2.3, HYPOXIA RESPONSIVE1 (HRE1) and HRE2, which are characterized by a conserved N-terminal motif (MCGGAII) (Nakano et al. 2006; Bailey-Serres et al. 2012). Four members of this group (except RAP2.3) have been reported to be associated with responses to submergence (flooding) and hypoxia (Papdi et al. 2008; Hinz et al. 2010; Licausi et al. 2010). Two recent studies have demonstrated that all five members in group VII ERFs could be degraded by the N-end rule pathway via a conserved N-terminal motif initiating with Met-Cys (Gibbs et al. 2011; Licausi et al. 2011). This represents a new mechanism of plant oxygen sensing regulated by the N-end rule pathway (Gibbs et al. 2011; Licausi et al. 2011). AtACBP1 and AtACBP2 were shown to participate in oxygen sensing through their interactions with RAP2.12 at the plasma membrane (Licausi et al. 2011). Under aerobic conditions, RAP2.12 was observed to be localized to the plasma membrane as indicated by fluorescent signals of the RAP2.12::GFP fusion (Licausi et al. 2011). Given the absence of a hydrophobic domain in RAP2.12, its membrane association suggests that there could be binding partners that sequester RAP2.12 at the plasma membrane. Considering the previous findings that AtACBP2 and AtACBP4 bind RAP2.3, two plasma membrane localized AtACBPs, AtACBP1 and AtACBP2 were checked whether they were the potential binding partners of RAP2.12 using BiFC and yeast two-hybrid assays (Licausi et al. 2011). Using a deletion mutant of RAP2.12, their results showed that RAP2.12 interacts with AtACBP1/AtACBP2 at the RAYD motif of RAP2.12 (Licausi et al. 2011), a domain that has been previously reported to mediate protein–protein interactions (Okamuro et al. 1997). These interactions between AtACBP1/AtACBP2 and RAP2.12 are important because their binding forms a membrane-bound complex as indicated by BiFC assays, thereby limiting the access of RAP2.12 to the nucleus (Licausi et al. 2011) and protecting RAP2.12 from degradation by the N-end rule pathway (Fig. 3) (Gibbs et al. 2011; Licausi et al. 2011). Interestingly, upon hypoxic conditions (i.e. flooding), membrane-bound RAP2.12 disappeared and RAP2.12 was observed in the nucleus to function in the activation of hypoxia-responsive gene expression (Fig. 3) (Licausi et al. 2011). During reoxygenation, RAP2.12 is degraded rapidly (within one hour) by proteolysis to deactivate the hypoxic response (Fig. 3) (Licausi et al. 2011).
According to the results from yeast two-hybrid screening, AtACBP1 also interacts with another TF, ABA-RESPONSIVE ELEMENT BINDING PROTEIN1 (AREB1) (Tse 2005), which is a transcriptional activator regulating ABA signaling in seed germination, seedling development and drought stress (Fujita et al. 2005; Yoshida et al. 2010). The expression of AREB1 was observed to be up-regulated in AtACBP1-overexpressors in response to ABA treatment (Du et al. 2013a), suggesting a positive role of AtACBP1 in ABA signaling.
Conclusion and perspective
Over a dozen protein partners of AtACBPs have been isolated the past decade (Li and Chye 2004; Li et al. 2008; Tse 2005; Gao 2009; Gao et al. 2009, 2010; Licausi et al. 2011; Du et al. 2013a). Investigations on these interactors have revealed novel roles of AtACBPs in development and stress responses. The interactions between AtACBP1/AtACBP2 and TFs were unexpected given their final protein destinations. However, this is a good model on how TFs can be sequestered from entering the nucleus to participate in the regulation of gene expression (Licausi et al. 2011). Although the newly identified mechanism of oxygen sensing in plants is significant and AtACBPs have been shown to play important roles in this process (Licausi et al. 2011; Bailey-Serres et al. 2012), a number of questions remain. The interaction between AtACBP1/AtACBP2 and RAP2.12 could be crucial in the hypoxic response (Licausi et al. 2011). Thus, it would be interesting to test whether the anoxia response and flooding tolerance fluctuate according to differential levels in AtACBP1/AtACBP2 expression. Membrane association of RAP2.12 was observed under normoxia but disappeared upon hypoxia (Licausi et al. 2011). Are the interactions between AtACBP1/AtACBP2 and RAP2.12 affected by oxygen levels? Is RAP2.12 released from membrane-bound AtACBP1/AtACBP2 upon hypoxia? Does RAP2.12 migrate from the plasma membrane (or cytosol) to the nucleus upon hypoxia? Are the cytosolic AtACBP4/AtACBP5 involved in the TF transfer given that AtACBP4 binds RAP2.3 (Li et al. 2008)? Does the binding of RAP2.12 to AtACBP1/AtACBP2 (also RAP2.3 to AtACBP2/AtACBP4) actually prevent degradation by the N-end rule pathway? If it does, would it affect E3 ligase recognition? Besides RAP2.12 and RAP2.3, do the other members in the group VII ERFs interact with AtACBPs at the plasma membrane? Previous studies have shown that AtACBP2 and AtACBP4 interact with RAP2.3 at the plasma membrane and cytosol, respectively (Li and Chye 2004; Li et al. 2008). AtACBP4 was observed to be localized to the periphery of nucleus likely arising from its interaction with RAP2.3 (Li et al. 2008). Considering that RAP2.3 can be degraded by the N-end rule pathway (Gibbs et al. 2011), does RAP2.3 also participate in the hypoxia response mimicking the oxygen-sensing mechanism of RAP2.12?
AtACBP1 has been observed to interact with AREB1 (Tse 2005) to promote ABA signaling in germination and vegetative growth (Du et al. 2013a). However, the mechanism of the AtACBP1 and AREB1 interaction is still unclear. AtACBP2 is involved in Cd(II) and H2O2 tolerance by the interaction with FARNESYLATED PROTEIN6 (AtFP6) (Gao et al. 2009), which is also known as the HEAVY METAL ASSOCIATED ISOPRENYLATED PLANT PROTEIN26 (HIPP26) (Barth et al. 2009). Although GFP::AtFP6 was co-localized with DsRed::AtACBP2 to the plasma membrane (Gao et al. 2009), AtFP6/HIPP26 was detected in the nucleus and found to interact with the stress-related ZINC FINGER HOMEODOMAIN TRANSCRIPTION FACTOR ATHB29 (Barth et al. 2009), indicating an indirect association between AtACBP2 and ATHB29. Furthermore, the results of yeast two-hybrid screens show a number of uncharacterized interactors of AtACBP1/AtACBP2 (Tse 2005; Gao 2009). The current knowledge on AtACBP interactors provides a good basis for further investigations to better understand the function of Arabidopsis ACBPs and their interactors.
Abbreviations
- ACBP:
-
Acyl-CoA-binding protein
- ACB:
-
Acyl-CoA-binding domain
- TF:
-
Transcription factor
- ERF:
-
Ethylene response factor
References
Adams J, Kelso R, Cooley L (2000) The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol 10:17–24
Andrade MA, Gonzalez-Guzman M, Serrano R, Rodriguez PL (2001) A combination of the F-box motif and kelch repeats defines a large Arabidopsis family of F-box proteins. Plant Mol Biol 46:603–614
Bailey-Serres J, Fukao T, Gibbs DJ, Holdsworth MJ, Lee SC, Licausi F, Perata P, Voesenek LA, van Dongen JT (2012) Making sense of low oxygen sensing. Trends Plant Sci 17:129–138
Barth O, Vogt S, Uhlemann R, Zschiesche W, Humbeck K (2009) Stress induced and nuclear localized HIPP26 from Arabidopsis thaliana interacts via its heavy metal associated domain with the drought stress related zinc finger transcription factor ATHB29. Plant Mol Biol 69:213–226
Burton M, Rose TM, Faergeman NJ, Knudsen J (2005) Evolution of the acyl-CoA binding protein (ACBP). Biochem J 392:299–307
Chen QF, Xiao S, Chye ML (2008) Overexpression of the Arabidopsis 10-kilodalton acyl-Coenzyme A-binding protein ACBP6 enhances freezing tolerance. Plant Physiol 148:304–315
Chen QF, Xiao S, Qi W, Mishra G, Ma J, Wang M, Chye ML (2010) The Arabidopsis acbp1acbp2 double mutant lacking the acyl-CoA-binding proteins ACBP1 and ACBP2 is embryo lethal. New Phytol 186:843–855
Chye ML (1998) Arabidopsis cDNA encoding a membrane-associated protein with an acyl-CoA binding domain. Plant Mol Biol 38:827–838
Chye ML, Huang BQ, Zee SY (1999) Isolation of a gene encoding Arabidopsis membrane-associated acyl-CoA binding protein and immunolocalization of its gene product. Plant J 18:205–214
Chye ML, Li HY, Yung MH (2000) Single amino acid substitutions at the acyl-CoA-binding domain interrupt 14[C]palmitoyl-CoA binding of ACBP2, an Arabidopsis acyl-CoA-binding protein with ankyrin repeats. Plant Mol Biol 44:711–721
Du ZY, Xiao S, Chen QF, Chye ML (2010) Depletion of the membrane-associated acyl-CoA-binding protein ACBP1 enhances the ability of cold acclimation in Arabidopsis. Plant Physiol 152:1585–1597
Du ZY, Chen MX, Chen QF, Xiao S, Chye ML (2013a) Arabidopsis acyl-CoA-binding protein ACBP1 participates in abscisic acid-mediated seed germination and seedling development. Plant J. doi:10.1111/tpj.12121
Du ZY, Chen MX, Chen QF, Xiao S, Chye ML (2013b) Overexpression of Arabidopsis acyl-CoA-binding protein ACBP2 enhances drought tolerance. Plant Cell Environ 36:300–314
Faergeman NJ, Feddersen S, Christiansen JK, Larsen MK, Schneiter R, Ungermann C, Mutenda K, Roepstorff P, Knudsen J (2004) Acyl-CoA-binding protein, Acb1p, is required for normal vacuole function and ceramide synthesis in Saccharomyces cerevisiae. Biochem J 380:907–918
Fan L, Zheng S, Cui D, Wang X (1999) Subcellular distribution and tissue expression of phospholipase Dα, Dβ, and Dγ in Arabidopsis. Plant Physiol 119:1371–1378
Feddersen S, Neergaard TB, Knudsen J, Faergeman NJ (2007) Transcriptional regulation of phospholipid biosynthesis is linked to fatty acid metabolism by an acyl-CoA-binding-protein-dependent mechanism in Saccharomyces cerevisiae. Biochem J 407:219–230
Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17:3470–3488
Gaigg B, Neergaard TB, Schneiter R, Hansen JK, Faergeman NJ, Jensen NA, Andersen JR, Friis J, Sandhoff R, Schroder HD, Knudsen J (2001) Depletion of acyl-coenzyme A-binding protein affects sphingolipid synthesis and causes vesicle accumulation and membrane defects in Saccharomyces cerevisiae. Mol Biol Cell 12:1147–1160
Gao W (2009) Characterization of protein interactors of Arabidopsis acyl-coenzyme a-binding protein 2. Dissertation, University of Hong Kong
Gao W, Xiao S, Li HY, Tsao SW, Chye ML (2009) Arabidopsis thaliana acyl-CoA-binding protein ACBP2 interacts with a heavy-metal-binding protein ATFP6. New Phytol 181:89–102
Gao W, Li HY, Xiao S, Chye ML (2010) Acyl-CoA-binding protein 2 binds lysophospholipase 2 and lysoPC to promote tolerance to cadmium-induced oxidative stress in transgenic Arabidopsis. Plant J 62:989–1003
Gibbs DJ, Lee SC, Isa NM, Gramuglia S, Fukao T, Bassel GW, Correia CS, Corbineau F, Theodoulou FL, Bailey-Serres J, Holdsworth MJ (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479:415–418
Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11:1897–1909
Guidotti A, Forchetti CM, Corda MG, Konkel D, Bennett CD, Costa E (1983) Isolation, characterization, and purification to homogeneity of an endogenous polypeptide with agonistic action on benzodiazepine receptors. Proc Natl Acad Sci USA 80:3531–3535
Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, Dolferus R (2010) Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol 153:757–772
Huang H, Atshaves BP, Frolov A, Kier AB, Schroeder F (2005) Acyl-coenzyme A binding protein expression alters liver fatty acyl-coenzyme A metabolism. Biochemistry 44:10282–10297
Kannan L, Knudsen J, Jolly CA (2003) Aging and acyl-CoA binding protein alter mitochondrial glycerol-3-phosphate acyltransferase activity. Biochim Biophys Acta 1631:12–16
Knudsen J, Faergeman NJ, Skott H, Hummel R, Borsting C, Rose TM, Anderson JS, Hojrup P, Roepstorff P, Kristiansen K (1994) Yeast acyl-CoA-binding protein: acyl-CoA-binding affinity and effect on intracellular acyl-CoA pool size. Biochem J 302:479–485
Knudsen J, Neergaard TB, Gaigg B, Jensen MV, Hansen JK (2000) Role of acyl-CoA binding protein in acyl-CoA metabolism and acyl-CoA-mediated cell signaling. J Nutr 130:294S–298S
Leung KC, Li HY, Mishra G, Chye ML (2004) ACBP4 and ACBP5, novel Arabidopsis acyl-CoA-binding proteins with kelch motifs that bind oleoyl-CoA. Plant Mol Biol 55:297–309
Leung KC, Li HY, Xiao S, Tse MH, Chye ML (2006) Arabidopsis ACBP3 is an extracellularly targeted acyl-CoA-binding protein. Planta 223:871–881
Li HY, Chye ML (2004) Arabidopsis acyl-CoA binding protein ACBP2 interacts with an ethylene-response element binding protein AtEBP via its ankyrin repeats. Plant Mol Biol 54:233–243
Li J, Mahajan A, Tsai MD (2006) Ankyrin repeat: a unique motif mediating protein–protein interactions. Biochemistry 45:15168–15178
Li HY, Xiao S, Chye ML (2008) Ethylene- and pathogen-inducible Arabidopsis acyl-CoA-binding protein 4 interacts with an ethylene-responsive element binding protein. J Exp Bot 59:3997–4006
Li M, Hong Y, Wang X (2009) Phospholipase D- and phosphatidic acid-mediated signaling in plants. Biochim Biophys Acta 1791:927–935
Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P (2010) HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J 62:302–315
Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LACJ, Perata P, van Dongen JT (2011) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479:419–422
Mandrup S, Sorensen RV, Helledie T, Nohr J, Baldursson T, Gram C, Knudsen J, Kristiansen K (1998) Inhibition of 3T3-L1 adipocyte differentiation by expression of acyl-CoA-binding protein antisense RNA. J Biol Chem 273:23897–23903
Meng W, Su YCF, Saunders RMK, Chye ML (2011) The rice acyl-CoA-binding protein gene family: phylogeny, expression and functional analysis. New Phytol 189:1170–1184
Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25:295–303
Michaely P, Bennett V (1992) The ANK repeat: a ubiquitous motif involved in macromolecular recognition. Trends Cell Biol 2:127–129
Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140:411–432
Oikari S, Ahtialansaari T, Heinonen MV, Mauriala T, Auriola S, Kiehne K, Folsch UR, Janne J, Alhonen L, Herzig KH (2008) Downregulation of PPARs and SREBP by acyl-CoA-binding protein overexpression in transgenic rats. Pflugers Arch Eur J Physiol 456:369–377
Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA 94:7076–7081
Papdi C, Abraham E, Joseph MP, Popescu C, Koncz C, Szabados L (2008) Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol 147:528–542
Rasmussen JT, Rosendal J, Knudsen J (1993) Interaction of acyl-CoA binding protein (ACBP) on processes for which acyl-CoA is a substrate, product or inhibitor. Biochem J 292:907–913
Rasmussen JT, Færgeman NJ, Kristiansen K, Knudsen J (1994) Acyl-CoA binding protein (ACBP) can mediate intermembrane acyl-CoA transport and donate acyl-CoA for β-oxidation and glycerolipid synthesis. Biochem J 299:165–170
Ryu SB (2004) Phospholipid-derived signaling mediated by phospholipase A in plants. Trends Plant Sci 9:229–235
Schjerling CK, Hummel R, Hansen JK, Borsting C, Mikkelsen JM, Kristiansen K, Knudsen J (1996) Disruption of the gene encoding the acyl-CoA-binding protein (ACB1) perturbs acyl-CoA metabolism in Saccharomyces cerevisiae. J Biol Chem 271:22514–22521
Sedgwick SG, Smerdon SJ (1999) The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem Sci 24:311–316
Tse MH (2005) Investigations on recombinant Arabidopsis acyl-Coenzyme A binding protein 1. Dissertation, University of Hong Kong
Xiao S, Chye ML (2009) An Arabidopsis family of six acyl-CoA-binding proteins has three cytosolic members. Plant Physiol Biochem 47:479–484
Xiao S, Chye ML (2011a) New roles for acyl-CoA-binding proteins (ACBPs) in plant development, stress responses and lipid metabolism. Prog Lipid Res 50:141–151
Xiao S, Chye ML (2011b) Overexpression of Arabidopsis ACBP3 enhances NPR1-dependent plant resistance to Pseudomonas syringe pv tomato DC3000. Plant Physiol 156:2069–2081
Xiao S, Li HY, Zhang JP, Chan SW, Chye ML (2008) Arabidopsis acyl-CoA-binding proteins ACBP4 and ACBP5 are subcellularly localized to the cytosol and ACBP4 depletion affects membrane lipid composition. Plant Mol Biol 68:571–583
Xiao S, Gao W, Chen QF, Chan SW, Zheng SX, Ma J, Wang M, Welti R, Chye ML (2010) Overexpression of Arabidopsis acyl-CoA-binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell 22:1463–1482
Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61:672–685
Zhang W, Qin C, Zhao J, Wang X (2004) Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phophatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci USA 101:9508–9513
Acknowledgments
This work was supported by the Hong Kong Research Grants Council (HKU765511 M) and by the Wilson and Amelia Wong Endowment Fund.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Du, ZY., Chye, ML. Interactions between Arabidopsis acyl-CoA-binding proteins and their protein partners. Planta 238, 239–245 (2013). https://doi.org/10.1007/s00425-013-1904-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-013-1904-2