Abstract
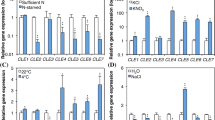
The cellulose synthase-like (ZmCSL) gene family of maize was annotated and its expression studied in the maize mesocotyl. A total of 28 full-length CSL genes and another 13 partial sequences were annotated; four are predicted to be pseudogenes. Maize has all of the CSL subfamilies that are present in rice, but the CSLC subfamily is expanded from 6 in rice to 12 in maize, and the CSLH subfamily might be reduced from 3 to 1. Unlike rice, maize has a gene in the CSLG subfamily, based on its sequence similarity to two genes annotated as CSLG in poplar. Light regulation of glycan synthase enzyme activities and CSL gene expression were analyzed in the mesocotyl. A Golgi-localized glucan synthase activity is reduced by ~50% 12 h after exposure to light. β-1,4-Mannan synthase activity is reduced even more strongly (>85%), whereas β-1,4-xylan synthase, callose synthase, and latent IDPase activity respond only slightly, if at all, to light. At least 17 of the CSL genes (42%) are expressed in the mesocotyl, of which four are up-regulated at least twofold, seven are down-regulated at least twofold, and six are not affected by light. The results contribute to our understanding of the structure of the CSL gene family in an important food and biofuel plant, show that a large percentage of the CSL genes are expressed in the specialized tissues of the mesocotyl, and demonstrate that members of the CSL gene family are differentially subject to photobiological regulation.







Similar content being viewed by others
Abbreviations
- CSL:
-
Cellulose synthase-like
- MLG:
-
Mixed-linked glucan
- HPAEC:
-
High performance anion exchange chromatography
- PAD:
-
Pulsed amperometric detector
References
Arioli T, Peng L, Betzner SA, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazenski J, Birch R, Cork A, Glover J, Redmond J, Williamson RE (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279:717–720
Barker-Bridgers M, Ribnicky DM, Cohen JD, Jones AM (1998) Red-light-regulated growth. Changes in the abundance of indole acetic acid in the maize (Zea mays L.) mesocotyl. Planta 204:207–211
Bernal AJ, Jensen JK, Harholt J, Sørensen S, Moller I, Blaukopf C, Johansen B, de Lotto R, Pauly M, Scheller H, Willats W (2007) Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J 52:791–802
Bernal AJ, Yoo C-M, Mutwil M, Jensen JK, Hou G, Blaukopf C, Sorensen I, Blancaflor EB, Scheller HV, Willats WGT (2008) Functional analysis of the cellulose synthase like genes CSLD1, CSLD2 and CSLD4 in tip growing Arabidopsis cells. Plant Physiol 148:1238–1253
Blaauw OH, Blaauw-Jansen G, Van Leeuwen WJ (1968) An irreversible red light-induced growth response in Avena. Planta 82:87–104
Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB (2006) Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1, 3;1, 4)-β-d-glucans. Science 311:1940–1942
Burton RA, Jobling SA, Harvey AJ, Shirley NJ, Mather DE, Bacic A, Fincher GB (2008) The genetics and transcriptional profiles of the cellulose synthase-like HvCSslF gene family in barley. Plant Physiol 146:1821–1833
Carpita NC (1996) Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol 47:445–476
Carpita NC, Defernez M, Findlay K, Wells B, Shoue DA, Catchpole G, Wilson RH, McCann MC (2001) Cell wall architecture of the elongating maize coleoptile. Plant Physiol 127:551–565
Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson C (2007) A gene from the cellulose synthase-like C family encodes a β-1, 4 glucan synthase. Proc Natl Acad Sci USA 104:8550–8555
Cona A, Cenci F, Cervelli M, Federico R, Mariottini P, Moreno S, Angelini R (2003) Polyamine oxidase, a hydrogen peroxide-producing enzyme, is up-regulated by light and down-regulated by auxin in the outer tissues of the maize mesocotyl. Plant Physiol 131:803–813
Cosgrove DJ (2005) Growth of the plant cell wall. Nat Rev Mol Cell biol 6:850–861
Dhugga KS, Barreiro R, Whitten B, Stecca K, Hazebroek J, Randhawa GS, Dolan M, Kinney AJ, Tomes D, Nichols S, Anderson P (2004) Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science 303:363–366
Doblin MS, De Melis L, Newbigin E, Bacic A, Read SM (2001) Pollen tubes of Nicotiana alata express two genes from different β-glucan synthase families. Plant Physiol 125:2040–2052
Dunkley TPJ, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, Watson RB, Dupree P, Lilley KS (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103:6518–6523
Farrokhi N, Burton RA, Brownfield L, Hrmova M, Wilson SM, Bacic A, Fincher GB (2006) Plant cell wall biosynthesis:genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotech J 4:145–167
Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L (2001) KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev 15:79–89
Fu Y, Emrich SJ, Guo L, Wen TJ, Ashlock DA, Aluru S, Schnable PS (2005) Quality assessment of maize assembled genomic islands (MAGIs) and large-scale experimental verification of predicted genes. Proc Natl Acad Sci USA 102:12282–12287
Gibeaut DM, Carpita NC (1991) Tracing cell wall biogenesis in intact cells and plants. Selective turnover and alteration of soluble and cell wall polysaccharides in grasses. Plant Physiol 97:551–561
Gibeaut DM, Pauly M, Bacic A, Fincher GB (2005) Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta 221:729–738
Goodwin RH (1942) On the development of xylary elements in the first internode of Avena in dark and light. Am J Bot 29:818–828
Goubet F, Misrahi A, Park SK, Zhang Z, Twell D, Dupree P (2003) AtCSLA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol 131:547–557
Hayashi T (1989) Xyloglucans in the primary cell wall. Annu Rev Plant Physiol 40:139–168
Hazen SP, Scott-Craig JS, Walton JD (2002) Cellulose synthase-like genes of rice. Plant Physiol 128:336–340. Available at http://waltonlab.prl.msu.edu/CSL_updates.htm
Hazen SP, Hawley RM, Davis GL, Henrissat B, Walton JD (2003) Quantitative trait loci and comparative genomics of cereal cell wall composition. Plant Physiol 132:263–271
Holland N, Holland D, Helentjaris T, Dhugga KS, Xoconostle-Cazares B, Delmer DP (2000) A comparative analysis of the plant cellulose synthase (CesA) gene family. Plant Physiol 123:1313–1324
Iino M, Carr DJ (1982) Sources of free IAA in the mesocotyl of etiolated maize seedlings. Plant Physiol 69:1109–1112
Inouhe M, Nevins DJ (1991) Auxin-enhanced glucan autohydrolysis in maize coleoptile cell walls. Plant Physiol 96:285–290
Jacobs A, Lipka V, Burton R, Panstruga R, Strizhov N, Schulze-Lefert P, Fincher G (2003) An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15:2503–2513
Jones AM, Cochran DS, Lamerson PM, Evans ML, Cohen JD (1991) Red light-regulated growth. Changes in the abundance of indoleacetic acid and a 22-kilodalton auxin-binding protein in the maize mesocotyl. Plant Physiol 97:352–358
Kim CM, Park SH, Je BI, Park SH, Park SJ, Piao HL, Eun MY, Dolan L, Han C (2007) OsCSLD1, a cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice. Plant Physiol 143:1220–1230
Li J, Burton RA, Harvey AJ, Hrmova M, Wardak AZ, Stone BA, Fincher GB (2003) Biochemical evidence linking a putative callose synthase gene with (1 → 3)-β-d-glucan biosynthesis in barley. Plant Mol Biol 53:213–225
Liepman AH, Wilkerson CG, Keegstra K (2005) Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA 102:2221–2226
Liepman AH, Nairn CJ, Willats WGT, Sørensen I, Roberts AW, Keegstra K (2007) Functional genomic analysis supports conservation of function among cellulose synthase-like A gene family members and suggests diverse roles of mannans in plants. Plant Physiol 143:1881–1893
Livak KJ, Schmittgen T (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Mitsui T, Honma M, Kondo T, Hashimoto N, Kimura S, Igaue I (1994) Structure and function of the Golgi complex in rice cells. II. Purification and characterization of Golgi membrane-bound nucleoside diphosphatase. Plant Physiol 106:119–125
Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 93:12637–12642
Pennisi E (2008) Corn genomics pops wide open. Science 319:1333
Piazza P, Jengkins ProcissiA, GI TonelliC (2002) Members of the c1/pl1 regulatory gene family mediate the response of maize aleurone and mesocotyl to different light qualitities and cytokinins. Plant Physiol 128:1077–1086
Pufky J, Qiu Y, Rao MV, Hurban P, Jones AM (2003) The auxin-induced transcriptome for etiolated Arabidopsis seedlings using a structure/function approach. Funct Integr Genomics 3:135–143
Ray PM, Shininger TL, Ray MM (1969) Isolation of β-glucan synthetase particles from plant cells and identification with Golgi membranes. Proc Natl Acad Sci USA 64:605–612
Richmond TA, Somerville CR (2001) Integrative approaches to determining Csl function. Plant Mol Biol 47:131–143
Roberts AW, Bushoven JT (2007) The cellulose synthase (CESA) gene superfamily of the moss Physcomitrella patens. Plant Mol Biol 63:207–219
Sawers RJ, Linley PJ, Gutierrez-Marcos JF, Delli-Bovi T, Farmer PR, Kohchi T, Terry MJ, Brutnell TP (2004) The Elm1 (ZmHy2) gene of maize encodes a phytochromobilin synthase. Plant Physiol 136:2771–2781
Saxena IM, Brown RM Jr, Fevre M, Geremia RA, Henrissat B (1995) Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J Bacteriol 177:1419–1424
Smith BG, Harris PJ (1999) The polysaccharide composition of Poales cell walls: Poaceae cell walls are not unique. Biochem System Ecol 27:33–53
Suzuki S, Li L, Sun YH, Chiang VL (2006) The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol 142:1233–1245
Taussky HH, Shorr E (1953) A microcolorimetric method for the determination of inorganic phosphorus. J Biol Chem 202:675–685
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 25:4876–4882
Tucker SC (1957) Ontogeny of the etiolated seedling mesocotyl of Zea mays. Bot Gaz 118:160–174
Walton JD, Ray PM (1982a) Inhibition by light of growth and Golgi-localized glucan synthase in the maize mesocotyl. Planta 156:302–308
Walton JD, Ray PM (1982b) Auxin controls Golgi-localized glucan synthase activity in the maize mesocotyl. Planta 156:309–313
Wang X, Cnops G, Vanderhaeghen R, De Block S, Van Montagu M, Van Lijsebettens M (2001) AtCSLD3, a cellulose synthase-like gene important for root hair growth in Arabidopsis. Plant Physiol 126:575–586
Yao H, Guo L, Fu Y, Borsuk LA, Wen TJ, Skibbe DS, Cui X, Scheffler BE, Cao J, Emrich SJ, Ashlock DA, Schnable PS (2005) Evaluation of five ab initio gene prediction programs for the discovery of maize genes. Plant Mol Biol 57:445–460
Yokoyama R, Rose JK, Nishitani K (2004) A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol 134:1088–1099
Zhu Y, Nam J, Carpita NC, Matthysse AG, Gelvin SB (2003) Agrobacterium-mediated root transformation is inhibited by mutation of an Arabidopsis cellulose synthase-like gene. Plant Physiol 133:1000–1010
Acknowledgments
This work was supported by grants from the National Science Foundation (DBI 0211797) and the US Department of Energy, Division of Energy Biosciences (DE-FG02-91ER20021). We thank members of Ken Keegstra and Curt Wilkerson’s laboratories for many helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
van Erp, H., Walton, J.D. Regulation of the cellulose synthase-like gene family by light in the maize mesocotyl. Planta 229, 885–897 (2009). https://doi.org/10.1007/s00425-008-0881-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-008-0881-3