Abstract
A long-standing mystery in the field of Polycomb and Trithorax regulation is how these proteins, which are highly conserved between flies and mammals, can regulate several hundred equally highly conserved target genes, but recognise these targets via cis-regulatory elements that appear to show no conservation in their DNA sequence. These elements, termed Polycomb/Trithorax response elements (PRE/TREs or PREs), are relatively well characterised in flies, but their mammalian counterparts have proved to be extremely difficult to identify. Recent progress in this endeavour has generated a wealth of data and raised several intriguing questions. Here, we ask why and to what extent mammalian PREs are so different to those of the fly. We review recent advances, evaluate current models and identify open questions in the quest for mammalian PREs.
Similar content being viewed by others
Introduction: the central mystery of fly and vertebrate PREs
The highly conserved Polycomb (PcG) and Trithorax (TrxG) group proteins work antagonistically on several hundred developmentally important target genes, to maintain repressed (PcG) or active (TrxG) transcription states (Simon and Kingston 2013; Steffen and Ringrose 2014). The repertoire of target genes that are regulated by the PcG and TrxG is remarkably similar in flies and vertebrates, including the Hox genes, many master transcriptional regulators, and genes involved in signalling and proliferation (Ringrose 2007). Given the identity of these targets, it is not surprising that aberrant expression of the PcG and TrxG proteins can lead to developmental defects and cancer (Kennison 2004; Richly et al. 2011). However, given the similarity of the target genes, together with the high conservation of the PcG and TrxG proteins themselves, it is extremely intriguing that the DNA sequences to which they bind show no apparent similarity between flies and vertebrates (Kassis and Brown 2013; Ringrose and Paro 2007).
In flies, the PcG and TrxG proteins act through Polycomb/Trithorax response elements (PREs). Whilst the properties of PREs and the DNA sequences that define them are reasonably well characterised in flies, the analogous elements in mammals have proved highly elusive (Kassis and Brown 2013; Steffen and Ringrose 2014). The effort to identify and understand mammalian PREs is currently one of the most active and controversial areas in the PcG/TrxG field.
Understanding the design principles and functions of mammalian PREs will be crucial for understanding genome-wide mammalian PcG/TrxG function in health and disease. Why are PREs not conserved? Do mammalian PREs use different sequences but perform essentially the same function as fly PREs? Or does mammalian PcG/TrxG regulation play by fundamentally different rules to those in the fly? In this review, we address these questions and evaluate recent progress in the quest for mammalian PREs.
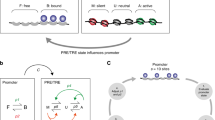
What makes a fly PRE? Different properties depend on context
PREs are best characterised in flies. These fascinating cis-regulatory elements work in concert with enhancers to ensure genome-wide transcriptional fidelity; however, PREs are distinct from enhancers in two key aspects. First, whereas enhancers respond to the cellular concentrations of transcription factors with exquisite precision in different cell types, PREs do not depend on the cellular concentrations of the PcG and TrxG proteins, which are ubiquitously expressed. Instead, PREs can adopt an active or silent state by responding to the status of their associated enhancers and promoters (Maeda and Karch 2006). Second, whereas enhancers can determine patterns of gene transcription, PREs alone cannot do so. However, PREs can maintain the transcriptional status that has initially been determined by transcription factors acting at enhancers. This maintenance can persist over many cell generations, even in the absence of the initial determining transcription factors (Chan et al. 1994). Thus, PREs can give stable epigenetic memory of both silenced and active transcriptional states (reviewed in Steffen and Ringrose 2014). However, despite this stability, PREs also have a built-in flexibility, allowing switching or modulation of their output in response to developmental, environmental or metabolic cues (Cavalli and Paro 1998; Herzog et al. 2014; Ost et al. 2014).
In summary, PREs as we know them from the fly can fulfil four tasks: first, they recruit PcG and TrxG proteins, and second, establish an active or silent state depending on inputs from their associated promoter and enhancer. Third, the PRE may maintain a memory of this state, and fourth, it may switch states upon new incoming signals. There is accumulating evidence that each of these four properties can be quantitatively different in different developmental contexts. For example, one PRE can be switched early in development but not later (Cavalli and Paro 1998), and another is biased towards activation in early development but prefers silencing at later stages (Herzog et al. 2014). Furthermore, it is becoming clear that different fly PREs have different inherent “personalities”. Just as some people have an excellent memory and others continuously forget things, different PREs have different abilities in each of the four tasks outlined above (Beuchle et al. 2001; Okulski et al. 2011). These differences are likely to be fundamentally important for the regulation of their target genes. Fly PREs are composed of multiple short DNA motifs, whose number and order is highly variable from one PRE to another (Ringrose and Paro 2007; Kassis and Brown 2013), and also varies for the same PRE across different fly species (Hauenschild et al. 2008). To fully understand the “PRE code”, it will be essential to understand how DNA sequence modulates PRE output via interaction of the PRE with PcG and TrxG proteins, the surrounding genomic landscape and incoming signals, to understand why different elements have different quantitative responses to specific inputs.
What makes a mammalian PRE? Do they have analogous functions to fly PREs?
Given this situation in the fly, where do we stand with mammalian PREs? The first fly PREs were discovered over 20 years ago (Chan et al. 1994; Kassis 1994; Simon et al. 1993), whereas the first mammalian PREs were described only 5 years ago (Sing et al. 2009; Woo et al. 2010). Since then, the vast majority of work in the mammalian field has focused on defining DNA elements that can fulfil the first task of PREs, namely to recruit PcG proteins (little attention has focused on TrxG recruitment). What is the evidence that mammalian PREs can or need to perform the other functions shown by fly PREs? To answer this question, we review similarities and differences between flies and vertebrates in the different components of this regulatory system, namely the PcG and TrxG proteins, their target genes, and the mammalian PREs defined so far. For each component, we ask whether these similarities and differences throw light on what makes a mammalian PRE.
We will not cover the recent large body of work on the involvement of 3D genome architecture in PcG and TrxG function, since large-scale spatial events occur downstream of the initial targeting of PcG and TrxG proteins to PREs. In addition, there is emerging evidence that these long-range interactions may be mediated by insulator elements rather than PREs themselves. These topics have been covered in detail in several recent reviews (Pirrotta and Li 2012; Noordermeer and Duboule 2013; Smigova et al. 2014; Cheutin and Cavalli 2014).
Fly and vertebrate PcG and TrxG proteins
PcG complexes: conserved at the core, with higher mammalian diversity
The core components of PcG complexes are remarkably conserved between flies and vertebrates (Fig. 1). However, the most striking difference is that vertebrate complexes make use of multiple alternative versions of some subunits, which can be deployed at different developmental stages, at different genomic locations, and can confer different properties on the complex (Gil and O’Loghlen 2014; Margueron and Reinberg 2011; Simon and Kingston 2013). This in turn may place different requirements on the PREs that recruit them.
Composition of PcG Complexes in flies and vertebrates. The Polycomb repressive complex 2 (PRC2) and Polycomb repressive complex 1 (PRC1) family of complexes are shown. Core subunits are shown in green for PRC2 and blue for PRC1. Alternate subunits, derived from multiple genes and if more than two, are shown in grey. Accessory proteins are shown in orange. Top: mouse complexes; bottom: Drosophila complexes. Selected histone modifications are shown: Red hexagons: histone H3 tail trimethylated at lysine 27 (H3K27me3); yellow ovals: histone H2A monoubiquitinated at lysine 119 (vertebrates) or 118 (fly). (H2AK119/118 Ub). a PRC2 consists of four core subunits, EZH1/2 (fly E(Z)), EED (fly ESC), SUZ12 (fly SU(Z)12), and RbAp46/48 (or RBBP7/4; fly NURF55) (Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002), and three accessory proteins, PCL (Walker et al. 2010), JARID2 (Herz et al. 2012; Kalb et al. 2014; Landeira et al. 2010; Li et al. 2010; Pasini et al. 2010a; Peng et al. 2009; Shen et al. 2009), and AEBP2 (Cao and Zhang 2004; Kalb et al. 2014). Alternate translation start site usage results in four different EED isoforms (not shown in the figure), which have different preferred histone substrates (Kuzmichev et al. 2004). PRC2 dimethylates and trimethylates histone H3 at Lys27 (H3K27me3) through the SET domain of EZH1/2 (fly E(Z)) (Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002). In addition, PRC2 can bind H3K27me3 via EED (Hansen et al. 2008; Margueron et al. 2009). b Canonical PRC1 consists of four core subunits, RING1A/B (fly dRING), CBX (fly PC), PCGF (fly PSC or SU(Z)2), and PHC (fly PH) (Gil and O’Loghlen 2014; Simon and Kingston 2009). PRC1 catalyses H2AK119Ub1 (in flies H2AK118Ub1) through its RING1A/B (fly dRING) subunit (Cao et al. 2005; de Napoles et al. 2004; Scheuermann et al. 2012; Wang et al. 2004a). Canonical PRC1 can bind H3K27me3 via the chromodomain of CBX2 or 7 (fly PC) (Bernstein et al. 2006b; Fischle et al. 2003; Min et al. 2003); however, different CBX proteins have different preferences for modified histone tails (Bernstein et al. 2006b), see main text and Fig. 2 for details. c Top: one class of vertebrate non-canonical PRC1s consists of three core subunits, RING1A/B, PCGF, and RYBP or YAF2 and various accessory proteins. The complexes are distinguished by different PCGF subunits. The complex containing PCGF1 (PRC1.1) also contains the histone H3K36 demethylase KDM2B. Other PCGF subunits copurify with other accessory proteins (orange) (Gao et al. 2012). Bottom: Drosophila dRAF is the most similar to vertebrate PRC1.1 and consists of dRING, PSC, and the histone H3K36 demethylase dKDM2 (Lagarou et al. 2008). Further non-canonical PRC1s exist and are reviewed in Gil and O’Loghlen (2014) and Simon and Kingston (2013). See main text and Table 1 for detail on molecular properties
Two key PcG complexes are Polycomb repressive complex 2 (PRC2, Fig. 1a) and Polycomb repressive complex 1 (PRC1, Fig. 1b). Both at the amino acid sequence level and at the level of subunit diversity, PRC2 is more evolutionarily conserved than PRC1 (reviewed in Margueron and Reinberg 2011; Ringrose and Paro 2004; Schuettengruber et al. 2007). In fly PRC2, a single subunit, enhancer of zeste (E(Z)), is used throughout development, mediating dimethylation and trimethylation of histone H3 on lysine 27 (H3K27me2/3) (Czermin et al. 2002; Muller et al. 2002). In contrast, in mammalian PRC2, this role is taken by the EZH2 or EZH1 subunit. These two closely related proteins have markedly different activities and expression patterns. EZH2 has similar catalytic activity to fly E(Z) (Cao et al. 2002; Kuzmichev et al. 2002) and is predominantly found in embryonic stem cells (ESCs) and proliferating cells, whereas EZH1 replaces EZH2 in specific differentiating and non-dividing cell types (Margueron et al. 2008; Shen et al. 2008; Stojic et al. 2011). The enzymatic activity of EZH1 appears to be context dependent: it has been reported to show similar activity to EZH2 in vitro (Shen et al. 2008), to have reduced activity in vivo (Margueron et al. 2008), and in some cases to promote transcriptional activation (Mousavi et al. 2012; Xu et al. 2015). Whether the enzymatic activity of fly E(Z) is modulated in specific cell lineages or at specific target genes to mirror the situation in vertebrates is not known.
In contrast to PRC2, vertebrate PRC1 comes in multiple flavours (Gil and O’Loghlen 2014). Each of the four core subunits in canonical PRC1 has between two and five versions (Fig. 1b). Some of these have overlapping functions, for example, RING1A and RING1B, which catalyse ubiquitination of histone H2A, can compensate for each other (de Napoles et al. 2004). However, other subunits may confer unique properties on the complex, for example, CBX7 is the primary ortholog present in PRC1 in ESCs, is required for the maintenance of pluripotency, and is downregulated upon differentiation. CBX2, CBX4 and CBX8 are directly repressed by CBX7 and are upregulated upon lineage commitment (Morey et al. 2012; O’Loghlen et al. 2012). Intriguingly, transient recruitment of PRC1 containing CBX8 is required for the transcriptional activation of several differentiation genes (Creppe et al. 2014). In molecular terms, these differences may in part be conferred by the different affinities of the CBX chromodomains for different modified histones (discussed in detail below (Bernstein et al. 2006b)). However, a recent study suggests that these different functions may not be relevant in some lineages (Pemberton et al. 2014). The authors studied genome-wide distribution of PcG orthologs CBX6, CBX7, CBX8, RING1 and RING2 in human fibroblasts, showing that these proteins colocalise at multiple sites; thus, their functions may be redundant rather than protein specific. In the fly, a single subunit (PC) takes the place of CBX in PRC1 throughout development (Simon and Kingston 2013). Again, it may well be that in the fly, the properties rather than the identities of the PC subunit are developmentally regulated, for example, by posttranslational modifications (Niessen et al. 2009). The fly SU(Z)2 protein shares homology with PSC, fulfils similar functions in in vitro assays (Lo et al. 2009) and coimmunoprecipitates with PC when PC is overexpressed (Poux et al. 2001); thus, it may also participate in PRC1 in vivo and modulate its function; however, this has not been addressed in detail.
Finally, in both flies and vertebrates, a class of non-canonical PRC1s has been identified, which lack CBX or PC proteins and contain the ubiquitin ligase RING (Fig. 1c) (Farcas et al. 2012; Gao et al. 2012; Lagarou et al. 2008; Sanchez et al. 2007). The vertebrate complexes contain additional subunits and are distinguished by different PCGF orthologs (see Fig. 1c). The complex containing PCGF1 (PRC1.1) also contains the histone H3K36 demethylase KDM2B, whereas other PCGF subunits copurify with other accessory proteins (Gao et al. 2012). Interestingly, PRC1s containing each of these different PCGFs have distinct genomic localisations and enzymatic activities compared to canonical complexes, indicating that each PCGF, or the accessory proteins associated with it, may drive targeting by different mechanisms (Gao et al. 2012). Drosophila dRAF is the most similar to vertebrate PRC1.1 and consists of dRING, PSC and the histone H3K36 demethylase dKDM2 (Lagarou et al. 2008).
What do these differences in complex diversity tell us about the tasks of PREs in flies and vertebrates? The increased combinatorial potential of mammalian PRC1 compared to its fly counterparts suggests that mammalian PREs would need to be able to contend with a much larger number of unique complexes, with varying subunit compositions and properties during development, potentially requiring different recruitment mechanisms. The fact that vertebrate complexes with different subunit compositions are distributed differently across the genome, and are developmentally regulated, strongly suggests that they may have different preferences for the underlying DNA sequences. Once recruited to a given PRE, the differences in enzymatic properties of various vertebrate complexes may have a profound effect on quantitative properties of the PRE, such as the stability of silencing and the ability to switch between active and silent states.
Molecular mechanisms of activation and silencing: highly conserved, with a few striking exceptions
The molecular mechanisms of PcG/TrxG-mediated silencing and activation have been covered in detail by several recent reviews (Kingston and Tamkun 2014; Lanzuolo and Orlando 2012; Simon and Kingston 2013). To inform our discussion of how mammalian PREs might compare to those of the fly, we present a parallel analysis of the molecular properties of the vertebrate and fly proteins (Table 1). With few exceptions, most of the molecular properties of PcG and TrxG proteins have indeed been demonstrated for both the fly and the vertebrate counterparts.
Interestingly, the molecular activities of the PcG and TrxG proteins fall into three main groups, listed separately in Table 1. The first group (Table 1(A)) contains activities of the PcG proteins that tend to reinforce each other and contribute to transcriptional silencing. For example, PRC1 catalyses the monoubiquitination of histone H2A, which can then be bound by PRC2, stimulating its activity towards methylating H3K27, which can in turn recruit both PRC2 and PRC1 (Table 1(A), rows 1–5). The second group of molecular activities are those of the activating TrxG proteins (Table 1(B)), and again, it is becoming clear that many of these activities cooperate and reinforce each other. Thus, for example, the TRX protein recruits the histone acetyltransferase CBP, which acetylates lysine 27 on histone H3 (among other residues), creating a binding platform for the TrxG protein BRD4, which in turn can phosphorylate RNA Polymerase II, converting it to the elongating form (Table 1(B), rows 4–7). Finally, the third group of activities are those in which a PcG-mediated activity directly antagonises that of a TrxG protein, and vice versa (Table 1(C)). Thus, for example, a lysine demethylase associated with a PcG complex removes the methylation at H3K36, which is catalysed by a TrxG protein, and at the same time promotes histone H2A ubiquitination by an associated PcG protein (Fig. 1c, Table 1(C), row 1).
What does this tell us about the properties of the system and how it may work at PREs? Together, these three groups of activities paint a picture of a bistable system, in which intermediate states are unstable, but once an impulse towards activation or silencing has begun, the system has an inherent molecular momentum that will tend to push it stably towards one or other state. Although several of these mechanisms have been elucidated in vitro and their in vivo relevance remains to be determined, the important feature for our discussion of fly versus mammalian PREs is that these properties appear remarkably conserved. Indeed, the similarity in function is highlighted by several studies showing that mouse PcG proteins can substitute for those of the fly in vivo (Atchison et al. 2003; Laible et al. 1997; Muller et al. 1995).
However, a closer look reveals several intriguing differences between flies and vertebrates that may be fundamentally important for determining the quantitative properties of the system. For example, PcG proteins carry multiple posttranslational modifications, many of which are on non-conserved residues (Kaneko et al. 2010; Niessen et al. 2009). These in turn offer opportunities for regulation, suggesting that quantitative properties of specific proteins (for example, enzymatic activities or binding affinities) may be differently regulated in flies and vertebrates. Two further examples are illustrated in Fig. 2. The PRC1 complex can compact chromatin, thus limiting access to remodelling factors and preventing transcriptional activation (see Table 1, row 9). A recent study (Grau et al. 2011) reports the intriguing finding that different proteins within the mouse and fly PRC1 complexes are responsible for mediating this compaction (Fig. 2a); thus, the extent of compaction may be quantitatively different in vivo and may again be subject to very different regulation via posttranslational modifications.
Evidence for different molecular mechanisms mediated by Drosophila and vertebrate PcG proteins. a Different proteins of the PRC1 complex mediate chromatin compaction in Drosophila and mouse (Grau et al. 2011). Purified PRC1 (see Fig. 1) from both fly and mouse can compact nucleosomal arrays in vitro; however, a different protein mediates this activity in the two species. Fly and mouse homologs of the proteins involved are shown. Red regions show domains required for compaction in each case, which are overrepresented in basic amino acids. Other domains and degree of conservation between mouse and Drosophila are indicated. b Alignment of the chromodomains of Drosophila Polycomb (PC, amino acids 15–77) and five mouse homologs (CBX, amino acids 1–62) redrawn from Bernstein et al. (2006b) and coloured according to the ClustalX colour scheme http://www.jalview.org/help/html/colourSchemes/clustal.html. On the right of the alignment, in vitro binding preferences of the different chromodomains from Bernstein et al. (2006b) are shown. Histone binding was addressed using modified peptides, Kds ranged between 12 and 49 μM. RNA binding was non-sequence specific. RNA-binding activity of the Drosophila PC chromodomain has not been reported to our knowledge
Another important example of quantitative differences between mouse and fly proteins is in the affinity of the chromodomain of the Polycomb protein (Pc, CBX in mammals) for different modified histone H3 tails (Fig. 2b). Although the preference of the fly PC chromodomain for H3K27me3 over H3K9me3 (Fischle et al. 2003; Min et al. 2003) is often assumed to be a major driving force for targeting, the vertebrate CBX proteins show no such preference in vitro (Bernstein et al. 2006b). Indeed, several CBX chromodomains bind equally well to both H3K9me3 and H3K27me3, and one (CBX4) shows a preference for H3K9me3 (Bernstein et al. 2006b) (Fig. 2b). Several, but not all, CBX chromodomains also bind to RNA (Bernstein et al. 2006b; Yap et al. 2010). The choice of CBX subunit for inclusion in PRC1 has a profound effect on the properties of the complex in vivo (Bernstein et al. 2006b; Creppe et al. 2014; O’Loghlen et al. 2012), and these differences may in part be mediated by the different properties of CBX chromodomains.
In summary, the qualitative properties of the system in terms of activation, silencing and switching appear to exist in flies and vertebrates. However, the examples discussed above show that quantitative parameters may be very differently regulated, potentially giving a different output of the system in terms of its effect on transcriptional regulation in specific cases. The extent to which these activities and their regulators are recruited to specific sites will depend in turn on the properties of the PRE.
Fly and vertebrate PcG target genes: Hox regulation is fundamentally different
The fly and mammalian genomes share several hundred PcG targets genes in common (reviewed in Ringrose 2007). Does similar gene identity and function imply a similar role for PcG proteins in their regulation, and thus similar tasks for their PREs? The best-characterised targets of PcG regulation in both flies and vertebrates are the Hox genes, which specify the identity of segments along the anterior-posterior axis of the developing embryo. In bilateral animals, including flies and vertebrates, the linear arrangement of Hox genes in the Hox complexes corresponds to the pattern in which they are expressed along the body axis of the animal, a phenomenon known as colinearity (Duboule 2007; Duboule and Morata 1994). Because of these striking similarities, one might expect that the PREs of the Hox genes perform analogous functions in flies and vertebrates. However, as pointed out and discussed in detail by Duboule (2007), several common assumptions about the similarity of fly and vertebrate Hox complexes are in fact erroneous. Here, we consider the implications of these differences for the Hox PREs.
The first striking difference is the relative size of the complexes in flies and vertebrates (Duboule 2007). Figure 3a shows the fly Hox complexes (ANT-C and BX-C) drawn to scale above the mouse HoxD complex, which is far more compact (about 6-fold shorter). It is immediately clear that although the relative order of paralogous genes is conserved, the space occupied both by the transcription units and by intergenic DNA is far greater in the fly than in the mouse. This has implications not only for the number of PREs that can exist within a given regulatory region (see light bars and arrowheads on fly complexes) but also for the distance over which they must communicate with their associated gene promoter.
Similarities and differences in Drosophila and vertebrate Hox gene regulation. a The Drosophila Antennapedia (ANT-C) and Bithorax (BX-C) complexes and the mouse HoxD complex are drawn approximately to scale, based on Duboule (2007) and Maeda and Karch (2009). Dark bars indicate exons (introns not shown for HoxD due to scaling); light bars and vertical arrowheads in ANT-C and BX-C indicate experimentally verified PREs (Ringrose and Paro 2004 and references therein). Genes and regulatory regions with a common colour are most closely related in sequence between fly and mouse, and thus belong to the same paralogy group (Duboule 2007). Note that the colour coding is not intended to reflect the different regulatory regions of ANT-C and BX-C as in Maeda and Karch (2009). b Pattern of histone H3 lysine 27 methylation at mouse HoxD (left) and fly BX-C (right) in specific tissues over developmental time. Left: summary of data from Soshnikova and Duboule (2009b). In embryonic stem cells, H3K27me3 covers the entire HoxD locus (top). In tail buds of E8.5 embryos (middle) and E9.5 embryos (bottom), Hox genes are sequentially activated leading to clearing of H3K27me3 from the locus. Right: summary of data from Bowman et al. (2014) and Maeda and Karch (2009). In early (0–2 h) embryos (top), the BX-C very probably lacks H3K27me3 and PcG proteins, based on indirect evidence ((Orlando et al. 1998; Petruk et al. 2012); see main text for details). In parasegment 7 of stage 5 (2–3 h) embryos (middle), appropriate Hox genes are activated and repressed by the gap and pair rule gene products (Maeda and Karch 2009). In the same parasegment of later (post 5 h) embryos, repressed domains gain H3K27me3 (Bowman et al. 2014)
A further striking difference between mouse and fly development is in the timing of segmentation. In the mouse, segments are sequentially added from anterior to posterior of the developing embryo over a matter of days, accompanied by the sequential activation of Hox genes along the complex (Soshnikova and Duboule 2009a). In contrast, in the fly, all segments arise simultaneously within a few hours and the Hox genes are simultaneously activated or repressed by the products of the gap and pair rule genes within this short time window (Maeda and Karch 2009) (Fig. 3b).
This difference is accompanied by fundamental differences in the behaviour of the PcG proteins during these early stages of development (Fig. 3b). In early development, the mouse Hox genes are entirely covered with H3K27 methylation (Soshnikova and Duboule 2009b). Sequential activation of the Hox genes leads to sequential removal of H3K27 methylation, culminating in an appropriate pattern for each segment ((Lan et al. 2007; Mazzoni et al. 2013; Soshnikova and Duboule 2009b). In contrast, in the fly, the Hox complexes very likely begin life in a naive state: very little H3K27 methylation is detectable in early embryos before Hox expression (Petruk et al. 2012), and PcG and TrxG proteins first become robustly detectable on most Hox PREs after the first 2 h of embryogenesis (Orlando et al. 1998). Soon after the gene expression state has been set by activators and repressors, the domain in which Hox genes must be repressed in a given segment becomes covered with H3K27 methylation (Bowman et al. 2014). In both mouse and fly, the end result is a sharp boundary between active and silent domains of the complex, but the route by which this is achieved is very different, and may have important implications for the required properties of the PREs in each case.
The Hox genes are one example of PcG target genes that are well studied in flies and vertebrates. Whether other common target genes also show differences in their mode of PcG regulation remains to be seen.
Recruitment of PcG proteins to DNA: the fly and the mammal diverge
The DNA sequences underlying PcG- and TrxG-binding sites appear to show little similarity between flies and mammals. In Drosophila, PcG and TrxG proteins require specific DNA-binding proteins to target PREs (reviewed in Kassis and Brown 2013; Ringrose and Paro 2007). Several of their recognition motifs are well characterised and are conserved between Drosophila species, although the genomic position of PREs and their exact sequence composition is not, indicating that PREs evolve rapidly (Hauenschild et al. 2008) whilst maintaining similar target domains (Schuettengruber et al. 2014). This rapid evolution of PREs may partially explain why mammalian PREs have been elusive. In addition, the DNA-binding proteins involved in recruiting fly PcG and TrxG are only partially conserved in mammals (summarised in Tables 2 and S1) and their contribution in PcG recruitment is currently not fully understood (reviewed in Do sequence-specific DNA-binding proteins recruit mammalian PcG and TrxG proteins? section below). Recent progress has given rise to various alternative models for mammalian PRE design, invoking CpG islands, binding sites for alternative DNA-binding factors, and non-coding (nc) RNAs as potential components of PREs. We review the evidence for each of these models below.

Are CpG islands PREs?
The idea that CpG islands may in fact be the long-sought vertebrate PREs has recently gained momentum (Farcas et al. 2012; Klose et al. 2013; Ku et al. 2008; Lynch et al. 2012; Mendenhall et al. 2010; Tanay et al. 2007). CpG islands are 1–2-kb regions of elevated G+C content and high density of CpG dinucleotides compared to the rest of the genome. Over evolutionary time, methylation of CpG dinucleotides elsewhere in the genome leads to their eventual depletion, because methylated cytosine tends to mutate to thymine. However, the vast majority of CpG islands escape this DNA methylation, and thus maintain a high density of CpG dinucleotides compared to their relative depletion in the rest of the genome. (Deaton and Bird 2011). In flies, which have little or no DNA methylation, there is no such depletion of CpG dinucleotides. The mouse and human genomes each contain approximately 24,000 CpG islands, 50 % of which map to annotated promoters, with the remainder likely to coincide with unannotated sites of transcriptional initiation (Illingworth et al. 2010; Deaton and Bird 2011). In mouse ESCs, almost all CpG islands coincide with H3K4me3, regardless of their transcriptional status (Bernstein et al. 2006a; Thomson et al. 2010). H3K4 methylation is recruited to CpG islands through the joint action of the Cpf1 protein (Thomson et al. 2010), and the TrxG proteins MLL1 and MLL2, all of which have a ZF-CxxC domain, which binds specifically to unmethylated CpG dinucleotides (Denissov et al. 2014; Hu et al. 2013; Long et al. 2013a). In addition, numerous studies have observed PRC1 and PRC2 proteins and H3K27me3 at approximately 30 % of these H3K4me3 marked CpG islands in ESCs (Bernstein et al. 2006a; Ku et al. 2008). Thus, CpG islands can recruit both PcG and TrxG proteins. Are these “bivalent” CpG islands PREs?
Although the correlation of PcG binding with CpG islands is striking, it may to some extent be misleading. Approximately 70 % of annotated gene promoters have a CpG island (Fig. 4a), and many studies have focused on promoter-proximal PcG-binding sites; thus, a correlation of this kind could simply be a consequence of what is found at promoters. Indeed, genome-wide profiling in ESCs has revealed that approximately one quarter to one third of PRC1- and PRC2-binding sites do not map to annotated promoters (Dietrich et al. 2012; Peng et al. 2009) and many of these PcG-bound intergenic sites do not contain CpG islands that conform to computational detection criteria (Dietrich et al. 2012; Hekimoglu-Balkan et al. 2012). On the other hand, computational prediction of GpG islands has been questioned by several recent studies. Illingworth et al. (2010) mapped CpG islands experimentally by CxxC Affinity Purification plus deep sequencing (CAP-seq). The study identified many “orphan” CpG islands that do not map to annotated promoters, are unmethylated in many cell types and are not detected by prediction algorithms. A recent evolutionary study (Long et al. 2013b) compared DNA methylation status across seven vertebrate genomes and found a high conservation of unmethylated DNA at promoters despite varying GC content. Whether these non-methylated islands also correspond to sites of PcG binding was not addressed, but these studies show that the definition of what constitutes a CpG island in vivo is not trivial and raise the issue of whether we should be thinking in terms of “non-methylated islands” rather than “CpG islands”.
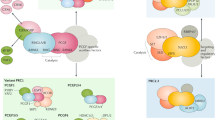
Recruitment of mammalian PcG complexes. a Relationship between occurrence of gene promoters, CpG islands, KDM2B and RING1B, according to Deaton and Bird (2011), Farcas et al. (2012), He et al. (2013), and Wu et al. (2013). b The RING1B subunit is a component of multiple different complexes, including both canonical and non-canonical PRC1 (Gao et al. 2012) see main text for details. c Factors influencing PcG recruitment. A stretch of GC- and CpG-rich DNA is shown (yellow). Various motifs for sequence-specific DNA-binding proteins can exist within this DNA (dark yellow), and several of these are themselves GC-rich (see Table 2). All of these motifs may also exist in otherwise GC-poor DNA. Proteins that can bind directly to DNA and have been shown or suggested to have role in PcG recruitment are shown in orange. PRC1: indicates all versions of PRC1 except the special case of PRC1.1 which is recruited by KDM2B. Arrows indicate that the DNA-binding protein in question does not copurify with the complexes but has been shown to interact by Co-IP. TA: activating transcription factor. See main text for details
Interestingly, in Xenopus embryos, sites of H3K27me3 nucleation in early development do not map to promoters, and no correlation between H3K27me3 domains and GC-rich sequences was observed in Xenopus or in zebra fish (van Heeringen et al. 2014). Importantly, a strong correlation was found between H3K27me3 and non-methylated DNA, rather than GC richness. Thus, PcG proteins can be recruited in the absence of strong CpG islands, and far from promoters, raising the question of whether the mammalian promoter sites recruit PcG proteins via the DNA sequences that define them as CpG islands, or whether there are other features of these sites such as their non-methylated status, or DNA sequence features other than GC-rich sites, that do the job. The Xenopus study (van Heeringen et al. 2014) strongly suggests that non-methylated status rather than CG richness per se may be essential. In addition, the frequency of CpG dinucleotides in computationally defined CpG islands is typically ten per 100 bases and the total GC content is 65 % (Wachter et al. 2014); thus, there is plenty of room for additional sequence features.
Indeed, several results argue against the necessity and sufficiency of CpG island-like features at the DNA sequence level for PcG recruitment. Several transgenic studies have demonstrated PcG recruitment to ectopic sites in the absence of a CpG island on the transgenic PRE (Schorderet et al. 2013; Sing et al. 2009; Woo et al. 2013). Xenopus sequences that recruit H3K27me3 in frog embryos and are GC-poor and unmethylated are able to repress a reporter and recruit H3K27me3 in mouse ESCs (van Heeringen et al. 2014). Furthermore, the deletion of sequences containing CpG islands from the endogenous mouse HoxD locus had no effect on the recruitment of H3K27me3 to flanking sites at the locus in vivo (Schorderet et al. 2013).
On the other hand, in support of a role for GC richness and CpG dinucleotides, several studies have shown that insertion of GC-rich sequences at ectopic sites is sufficient to recruit both H3K4me3 and H3K27me3 (Jermann et al. 2014; Lynch et al. 2012; Mendenhall et al. 2010; Wachter et al. 2014). However, below a certain threshold of C+G and CpG dinculeotides, these transgenic sequences become methylated and lose both H3K4me3 and H3K27me3 (Wachter et al. 2014) (Jermann et al. 2014). Further supporting the idea that PcG proteins will bind to GC-rich sites if not blocked by methylation, artificial reduction of endogenous DNA methylation has been shown to lead to a widespread redistribution of both PRC1 and PRC2 to GC-rich sites that were previously methylated (Brinkman et al. 2012; Cooper et al. 2014; Hagarman et al. 2013; Lynch et al. 2012; Reddington et al. 2013). In summary, the available evidence points towards several potential mechanisms of PcG recruitment, which may be complementary and are not mutually exclusive.
Recent mechanistic insights into one of these mechanisms have been provided by the identification of a direct link between one version of PRC1 and its localisation at unmethylated CpG islands. The H3K36 histone demethylase KDM2B (or FBXL10), part of a non-canonical PRC1 (Fig. 1c), specifically recognises non-methylated CpG dinucleotides through its ZF-CxxC domain, and this interaction is required to recruit some PRC1 proteins to a subset of CpG islands (Farcas et al. 2012; He et al. 2013; Long et al. 2013a; Wu et al. 2013), and to prevent DNA methylation (Boulard et al. 2015).
A closer look at the data raises important open questions (Fig. 4a) (Farcas et al. 2012; He et al. 2013; Wu et al. 2013). KDM2B was found to bind to all CpG islands genome-wide, whereas PRC1 components were found at only 15 to 30 % of these sites, raising the question of why PRC1 selects some sites and not others. Furthermore, the ChIP overlaps in Farcas et al. (2012), He et al. (2013) and Wu et al. (2013) were evaluated on the basis of the RING1B protein, which participates in a multitude of different canonical and non-canonical PRC1s, only one of which (PRC1.1) contains the KDM2B protein (Gao et al. 2012) (Figs. 1c and 4b). PRC1.1 binds to distinct sites from other PRC1s and is uniquely characterised by the presence of PCGF1 (and not other PCGFs) and the absence of CBX proteins (Gao et al. 2012) (Fig. 4b). An evaluation of the overlap of RING1B with these subunits would give insights into what proportion of the RING1B bound sites are directly recruited by KDM2B; however, these experiments were not performed in any of the above studies (Farcas et al. 2012; He et al. 2013; Wu et al. 2013). Thus, RING1B may be recruited to many of these sites independently of KDM2B, despite their coincident occurrence. The fact that KDM2B knockdown led to significant loss of RING1B at only 17 % of its targets (Wu et al. 2013) and upregulation of a small number of genes (78 genes of approximately 2000 RING1B bound targets (Farcas et al. 2012)) is consistent with this interpretation.
Nevertheless, these studies identify a direct DNA-based mechanism linking CpG dinucloetides to recruitment of PRC1 members. In future, it will be essential to determine the abundance and developmental regulation of various non-canonical complexes, to understand how they contribute to global targeting. The self-reinforcing nature of many PcG-based mechanisms raises the possibility that a transient recruitment by one complex may be sufficient to trigger a cascade of reactions leading to stable silencing (Blackledge et al. 2014) (Table 1), whose dynamic establishment escapes detection by ChIP profiling. Interestingly, the fly KDM2B homolog, dKDM2, also has a ZF-CxxC domain and participates in a non-canonical PRC1 (dRAF, see Fig. 1c, Tables 2 and S1). The fly genome is abundant in unmethylated CpG dinucleotides; however, the DNA-binding properties and genome-wide distribution of dKDM2 have not been evaluated.
Finally, the extremely intriguing question remains of why mammalian PcG proteins do not bind to all CpG islands, or all sites of KDM2B occupancy. The transgenic studies described above (Jermann et al. 2014; Wachter et al. 2014) clearly demonstrate that recruitment of bivalent chromatin is a default property of unmethylated CpG island-like sequences, raising the question of why all CpG islands do not recruit PcG in vivo, since they are unmethylated and share the sequence features that define CpG islands. Several models have been proposed to account for this discrepancy and fall into two broad, non-mutually exclusive classes. A “chromatin sampling” model proposes that PcG proteins weakly interact with all potential sites, but that transcriptional activity prevents PcG from stably binding potential sites (Klose et al. 2013). Thus, in this model, stable binding is only nucleated in response to a silent promoter, in a similar manner to the proposed mechanism in flies (reviewed in Steffen and Ringrose 2014). Indeed, the occurrence of activating transcription factor motifs or the placement of an active promoter at a CpG island have been shown to be sufficient to block the binding of PRC2 (Caputo et al. 2013; Jermann et al. 2014; Mendenhall et al. 2010; Riising et al. 2014), and global inhibition of transcription leads to a substantial invasion of these silenced sites by PRC2 (Riising et al. 2014). The idea that PcG targeting is merely a result of silencing at permissive CpG islands is neat and simple; however, there are also data that are inconsistent with this model: 30 to 40 % of CpG islands do not acquire PRC2 even after transcriptional inhibition (Riising et al. 2014), and conversely, 10–20 % of active genes do in fact show PcG occupancy is ESCs (reviewed in Ringrose 2007). There must be something more, so what is it?
Do sequence-specific DNA-binding proteins recruit mammalian PcG and TrxG proteins?
An alternative to this “responsive” model is an “instructive model”, which proposes that sequence-specific DNA-binding proteins recruit PcG and/or TrxG proteins to specific sites, as in the fly (reviewed in Klose et al. 2013). Binding sites for these factors may be embedded in CpG islands and may also occur elsewhere in the genome, giving an additional layer of specificity. These two models are not mutually exclusive, and we propose that their relative contributions at specific genomic sites and developmental stages will be different and that quantitative understanding of these contributions may hold the key to the vertebrate “PRE code”.
The DNA-binding proteins that have been shown to recruit PcG and TrxG proteins in flies and vertebrates are compared in Tables 2 and S1, and a selection is shown in Fig. 4c. The fly proteins PHO and GAF have mammalian homologs that may play a role in PcG regulation. However, the involvement of YY1 (Pho homolog) with PcG is debated. Whereas the mouse YY1 protein can rescue fly pho mutants and repress in flies, no silencing was observed in mammalian cell culture (Srinivasan et al. 2005; Wilkinson et al. 2006). Moreover, genome-wide mapping of PRC1/2 and YY1 showed few overlapping sites in ESCs, rather YY1 sites tend to colocalise with H3K4me3-enriched promoters (Mendenhall et al. 2010; Squazzo et al. 2006). Contrastingly, YY1 sites were required for repression mediated by two transgenic PREs (HoxD11.12 and HoxC11.12) but not at a third (HoxB4.5) (Woo et al. 2010, 2013). We note that since YY1 sites contain the motif “GCC”, their mutation may interfere with other proteins that bind GC-rich sequences (Tables 2 and S1). The mammalian homolog of GAGA factor (c-Krox-Th-POK or mGAF) has recently been identified and predicted to bind to the same DNA motif (GAGAG, Table 2) as in flies (Matharu et al. 2010). Profiling of mammalian Hox loci revealed intergenic binding sites for mGAF that are enriched for this motif (Srivastava et al. 2013), and the motif is enriched in several mammalian PREs (Fig. 5), but the involvement of mGAF in recruiting PcG proteins has not been investigated.
Motif occurrences in mammalian PREs. a A selection of mammalian PREs that have been verified to recruit PcG proteins in transgenic assays are shown (see Table 3 for details): HoxC11-12, HoxB4-5 (Woo et al. 2013), HoxD11-12 (Woo et al. 2010), PREkr (Sing et al. 2009), HoxD10 (Schorderet et al. 2013), DBE (Cabianca et al. 2012). Above each element, the % GC is shown, with CpG islands marked in dark grey, according to the following criteria: window size 100; minimum length of an island 200; minimum observed/expected CpG 0.6; minimum % GC 50.0. NB with these settings the HoxD10 PRE scores a short 200 bp GpG island; however, this was not detected by the more stringent settings used by Schorderet et al. (2013) and is designated as having no CpG island in Table 3 according to the authors of that study. Motifs for the DNA-binding proteins shown were scored as regular expressions with no mismatch allowance, as follows: REST: NTCAGCACCNNGGACAGCNCC; CP2: GCNCNANCCAG; RUNX:TGYGGT; YY1: GCCAT; GAF: GAGAGA, using the IUPAC code for non-conserved nucleotides as described in the legend to Table 2. b Occurrence per kb of motifs in the PREs shown and in random sequence (black). To generate random sequence, the total sequence of all elements shown (10.67 kb) was shuffled and searched for motifs. The mean of four iterations is shown
Several additional proteins have been identified in mammals that have a role in PcG targeting. The transcription factors REST, RUNX1 and E2F6 have been linked to PcG recruitment. E2F6 interacts with and colocalises with non-canonical PRC1s and may recruit these to specific sites (Ogawa et al. 2002; Trimarchi et al. 2001; Trojer et al. 2011). REST and RUNX1 coimmunoprecipitate with PcG proteins, and each occupy a subset of genomic loci bound by PRC1 members on a genome-wide scale. Five percent of REST-binding sites (Dietrich et al. 2012; Ren and Kerppola 2011) and 57 % of RUNX1-binding sites colocalise with PRC1 (Yu et al. 2012). Depletion of REST or RUNX1 leads to decreased PRC1 binding at common binding sites (Dietrich et al. 2012; Yu et al. 2012). Importantly, REST-mediated recruitment of PcG proteins is context dependent: PRC2 depends on REST for its recruitment to GC-rich sequences specifically in neural progenitors and not in ESCs (Arnold et al. 2013), whilst PRC1 can be recruited by REST independently of CpG islands in ESCs (Dietrich et al. 2012). These proteins have fly homologs (Tables 2 and S1), but their connection to PcG function has not been investigated.
In contrast to the above examples, the JARID2 protein is globally required for PRC2 recruitment (Landeira et al. 2010; Li et al. 2010; Pasini et al. 2010a; Peng et al. 2009). JARID2 copurifies and is highly colocalised genome-wide with PRC2 in ESCs (90 % overlap of ChIP-seq peaks (Landeira et al. 2010; Li et al. 2010; Pasini et al. 2010a; Peng et al. 2009). JARID2 can directly bind DNA in vitro without a clear sequence preference; thus, the DNA-binding activity alone is unlikely to add specificity (Tables 2 and S1). Indeed, although JARID2 is required for PRC2 recruitment, direct evidence that this occurs via DNA binding in vivo is lacking (Tables 2 and S1). The recent demonstration that PRC2 containing JARID2 and AEBP2 is able to bind to ubiquitinated histone H2A and that this enhances its HMTase activity, raises the possibility that recruitment involves recognition of existing H2Aub, giving another potential example of a self-reinforcing mechanism (Kalb et al. 2014). This in vitro interaction is conserved for the fly proteins (Kalb et al. 2014); however, fly Jarid2 mutants do not give Polycomb phenotypes (Sasai et al. 2007) and JARID2 does not colocalise highly with PRC2 in flies; thus, whether it has a role in targeting fly PRC2 is unclear (Herz et al. 2012). Consistent with a central role for JARID2 in modulating PRC2 activity, developmentally regulated methylation of mouse JARID2 by EZH2 also increases PRC2 HMTase activity (Sanulli et al. 2015).
Interestingly, mammalian REST, RUNX1, E2F6 and AEBP2 are expressed in specific tissues or developmental stages or at specific times during the cell cycle (Kherrouche et al. 2001) (see Tables 2 and S1). Thus, the recruitment mechanisms that depend on these factors may be required to give a boost of recruitment to relevant targets in specific contexts. This may imply that generally PcG proteins are recruited by other mechanisms. However, it could also imply that we have yet to identify the other DNA-binding factors that specifically target PcG complexes to different classes of targets. This would require either that PcG proteins have acquired specialised functions to recognise a variety of DNA-binding proteins, or that these proteins modulate the accessibility of chromatin to enable access for non-sequence-specific factors such as JARID2.
Do non-coding RNAs recruit PcG and TrxG proteins? A question of specificity
In Drosophila and vertebrates, many PcG target sites are transcribed into ncRNA (Brockdorff 2013; Hekimoglu and Ringrose 2009). This observation, in combination with the fact that several PcG and TrxG proteins can bind to RNA in vitro (Bernstein et al. 2006b; Krajewski et al. 2005), has given rise to the proposal that interactions of PcG and TrxG proteins with specific ncRNAs are responsible for targeting to specific sites in vivo (Cifuentes-Rojas et al. 2014; Kaneko et al. 2014; Zhao et al. 2008). Indeed, several ncRNAs have been shown to be required for PcG and TrxG function in vivo with exquisite specificity (reviewed in Brockdorff 2013; Grossniklaus and Paro 2014; Hekimoglu and Ringrose 2009). However, there are also results that argue against a simple targeting function for ncRNAs. The idea that PcG complexes themselves recognise specific RNA sequence or structural motifs is inconsistent with several observations. First, it is difficult to imagine how a generic protein complex can interact specifically with several hundred different RNA molecules in a pool of several thousand highly abundant other RNA species. Second, several studies have recently shown that PRC2 interacts promiscuously with RNA in vitro (Davidovich et al. 2013) and that this interaction leads to an inhibition of histone-methyltransferase activity of both fly and vertebrate PRC2 (Cifuentes-Rojas et al. 2014; Herzog et al. 2014). It remains to be seen whether other enzymatic activities of the PcG and TrxG complexes are stimulated or inhibited by RNA. A recent study examined Xist RNA using superresolution microscopy and observed that Xist RNA and PcG proteins do not in fact colocalise at high resolution, suggesting that Xist is unlikely to directly recruit PcG proteins to the inactive X-chromosome (Cerase et al. 2014).
A key future challenge will be to reconcile the lack of inherent specificity of PcG and TrxG proteins for specific RNAs in vitro, with the exquisite specificity of some ncRNAs in affecting PcG and TrxG function in vivo. We envisage three non-exclusive models. First, many of the ncRNAs arising from PcG-binding sites are highly developmentally regulated (Dinger et al. 2008; Guttman et al. 2011; Hekimoglu-Balkan et al. 2012; Wang et al. 2011; Herzog et al. 2014). This may offer a means to rapidly modulate PcG and TrxG enzymatic function without affecting recruitment, as observed by Herzog et al. (2014). Second, highly expressed transcripts could represent a decoy for PcG proteins to be displaced from the locus, as proposed by Davidovich et al. (2013) and observed by Herzog et al. (2014). Finally, there may be other specific binding factors that prevent or enable the interaction of certain RNAs with PcG and /or TrxG proteins, as proposed by Herzog et al. (2014). This would enable reversible and regulated switching of the availability of a given RNA to interact with PcG proteins. Although it remains to be seen to what extent these ideas are globally applicable, the important insight from recent work is that ncRNAs are unlikely to be involved in specific targeting, and rather to be involved in modulating the properties of PREs and the proteins bound there.
Experimental assays for fly and vertebrate PREs
How do we know that a PRE is a PRE? Although genome-wide profiling can give important insights into the localisation and interdependence of chromatin-binding proteins, transgenic reporter assays are essential for defining the DNA sequences that have PRE function.
Assays in fly
There are several transgenic assays for PRE function that are routinely used in flies. The most commonly used assay is the “miniwhite” reporter, in which transgenic flies are generated, carrying a candidate PRE linked to a minimal promoter driving a reporter gene (miniwhite). The miniwhite reporter is derived from the white gene, which gives a red eye colour in adult flies. Homozygous miniwhite transgenes typically give eye pigment levels of 15 to 50 % of that found in wild-type flies (Okulski et al. 2011). The repressive properties of the PRE, the effects of mutating different motifs and the response of the element to PcG and TrxG mutations are easily scored by quantification of eye pigment levels. The ability of the PRE to recruit PcG and TrxG proteins to the site of the transgene is evaluated by analysis of polytene chromosomes or by ChIP (reviewed in Ringrose and Paro 2004). In addition, fly PREs typically show pairing sensitive silencing (PSS) of miniwhite, whereby the reporter is more strongly silenced in homozygotes (carrying two copies of the transgene) than in heterozygotes (carrying a single copy; Kassis 1994). Recently, it has become possible to perform this assay by integrating different constructs at an identical genomic location, thus enabling a quantitative analysis of the effects of genomic position, to which fly PREs are extremely sensitive (Okulski et al. 2011). Several further assays that also address the ability of the PRE to preserve epigenetic memory of previously established transcriptional states in the absence of the initiating signal have also been devised, showing that several fly PREs can maintain memory of both active and silent states over many cell generations. These assays are more biologically relevant than the miniwhite assay as they address the maintenance of both silencing and activation over developmental time (Simon et al. 1993; Chan et al. 1994; Cavalli and Paro 1998; Perez et al. 2011). The strength of this memory function varies with both the genomic location, the identity of the PRE, and the developmental stage at which the signal to activate or silence is given, indicating that PRE properties are context dependent (reviewed in Steffen and Ringrose 2014).
Assays in mammalian systems
Several studies that have reported transgenic assays for mammalian PREs or PRE-like sequences are summarised in (Table 3). Comparison of these studies reveals that a wide range of contexts have been used. In contrast to the fly, very few studies have analysed PRE function in living animals (for example, Schorderet et al. 2013; Sing et al. 2009), instead the majority of studies rely on cell culture based assays, allowing rapid evaluation of candidate elements. Given the tractability of these systems, it is surprising that only 9 out of 15 studies assessed the ability of candidate PREs to repress a reporter. Each of these used a different reporter-promoter combination (Table 3). The other studies focused exclusively on the analysis of recruitment of PRC1 and/or PRC2 proteins to putative PREs at ectopic sites (Table 3). A variety of transiently transfected and integrated systems have been used (Table 3). Of the eight studies that used integrated transgenes, only four used targeted integration to compare different elements or variants at the same genomic location, which was a different site in three of the four studies (Arnold et al. 2013; Jermann et al. 2014; Lynch et al. 2012; Riising et al. 2014). Moreover, these studies cover eight different mammalian cell types, and several studies include Drosophila reporter assays (Basu et al. 2014; Bengani et al. 2013; Cuddapah et al. 2012; Sing et al. 2009; Vasanthi et al. 2013). Remarkably, studies assessing the binding of TrxG proteins are almost completely lacking. Finally, none of these studies addressed memory properties of mammalian PREs, in terms of testing whether the element can maintain a previously established silent or active state over cell generations in the absence of the initial determining signal. Thus, at present, although there are now a fairly large number of published elements that share some properties of fly PREs, in most cases, it is not possible to draw quantitative comparisons between different elements, and thus, it is very difficult to discern the connection between DNA sequence features and functional properties.
Computational analysis of fly and vertebrate PREs
An alternative for understanding the sequence principles of PREs is computational prediction. Do we still need prediction now that we have genome-wide profiling? Profiling technologies have revolutionised the way we address many questions in gene regulation, allowing the quantification of expression levels of hundreds of genes and the identification of thousands of protein-binding sites in the genome. However, in the search for mammalian PREs, genome-wide profiling has several caveats as shown in Fig. 6, because not every site of protein enrichment or histone modification is likely to be a PRE. Multiple ChIP peaks or large domains may represent both primary recruitment sites, and secondary sites to which the proteins spread or loop after recruitment by PREs. Furthermore, different tissues show different ChIP profiles and no single tissue will give information on all potential sites that can act as PREs.
Different modes of PcG binding and their resulting ChIP-binding profiles. On the left are shown different modes of dynamic binding of PcG proteins to PREs. On the right are shown the ChIP profiles that would result from each mode of binding. a Spreading. PcG proteins are recruited by a PRE and subsequently spread up and downstream (left), resulting in a broad ChIP peak (right) from which the PRE is not identifiable. b Looping. PcG proteins are recruited by a PRE and subsequently loop to the promoter via higher order interactions (left), resulting in two ChIP peaks (right) only one of which is a bona fide PRE. c Dynamic changes. In the example shown, PcG proteins are recruited by a PRE and are subsequently delivered to a different location (left), resulting in a ChIP peak at the site of delivery (in this example, the gene) but not at the site of entry (in this example, the PRE) (right). Variations on this theme include different profiles in different cell types, in which only a subset of multiple PREs may be occupied in different tissues or at different times
Computational prediction approaches can be used to complement profiling data, because they generate a model of underlying sequence determinants that can be highly valuable for distinguishing primary from secondary sites, and for identifying sequence characteristics that are not limited by tissue specificity. For example, computational prediction of fly PREs based on known motifs and trained to distinguish verified PREs from non-PREs identified approximately 20 % of ChIP-binding sites that were later observed in Drosophila embryos (Ringrose et al. 2003), discussed in (Hauenschild et al. 2008). Besides identifying many new PREs that were not known in 2003, comparison with later profiling datasets enables a classification into three categories: (i) predicted sites that are also ChIP enriched (these sites contain the motifs in question in particular configurations and are enriched in ChIP experiments); (ii) predicted sites that do not contain a ChIP enrichment in the tissue in question (these sites are computationally indistinguishable from class (i)); and (iii) non-predicted sites (those that have a ChIP enrichment but do not contain the motifs used for the prediction). This classification opens the door to formulating relevant questions: for example, why are some predicted sites bound and others not? Why are some bound sites not predicted? Indeed, inclusion of comparative genomic information from different fly species increases the overlap between ChIP and predictions to 34 % (Hauenschild et al. 2008), which is in the range of overlap observed between different ChIP-profiling studies of PcG proteins (28–34 % (Hauenschild et al. 2008) and references therein). Computational prediction of vertebrate PREs is less straightforward since it is currently less clear which DNA-binding proteins or DNA sequence features initiate PcG binding.
Another limitation is imposed by the small number of verified vertebrate PREs and the lack of specific DNA motifs therein. Interestingly, sequence mining of individual vertebrate PREs using fly motifs mainly identified GAF/mGAF and Pho/YY1 motifs (Bengani et al. 2013; Cabianca et al. 2012; Sing et al. 2009; Woo et al. 2010). However, these short motifs occur frequently in any sequence, and their general relevance to vertebrate PREs is not clear. Figure 5 shows an extended analysis of the occurrence of DNA motifs and sequence features discussed above (Do sequence-specific DNA-binding proteins recruit mammalian PcG and TrxG proteins? section) in a selection of verified mammalian PREs. This reveals that although several motifs do recur in vertebrate PREs (Fig. 5a), only the GAF motif occurs more frequently in PREs than in random sequence (Fig. 5b). Furthermore, there are large stretches of sequence in each PRE in which none of the motifs occur, so there may be additional motifs yet to be found. De novo motif discovery of PRC2 genome-wide binding sites further identified overrepresented repeat sequences, for example, GA and GC repeats (Hajjari et al. 2014; Hunkapiller et al. 2012; Kim et al. 2009; Peng et al. 2009). Cross-species analysis of intergenic and intronic Hox sequences revealed enrichments of GA repeats and poly-T stretches in comparison to housekeeping gene sequences (Bengani et al. 2007). Analysis of repetitive sequences in H3K27me3 domains in Xenopus revealed high enrichments (over 50-fold) of TAGA and TG repeats (van Heeringen et al. 2014). Distinct motifs have been identified in BMI1 (Meng et al. 2010) and AEPB2 peaks (Kim et al. 2009) (see Tables 2 and S1).
Going beyond motif discovery, several computational approaches have identified sequence features that predictively distinguish PcG-binding sites from non-target sites. As discussed above, several studies have shown a strong correlation between CpG islands and PcG binding (Hunkapiller et al. 2012; Ku et al. 2008; Mendenhall et al. 2010; Tanay et al. 2007) and further analysis identified binding sites for known transcription factors able to discern 2/3 of PcG-bound CpG islands from non-bound CpG islands in ESCs (Ku et al. 2008). Binding motifs for REST and SNAIL have been shown to be predictive for dynamic changes in H3K27me3 during neural differentiation (Arnold et al. 2013). The most thorough prediction to date trained a predictive algorithm to distinguish H3K27me3 enriched from non-enriched domains, on the basis of enrichment and depletion of 8-mer motifs (van Heeringen et al. 2014). The algorithm was initially trained on Xenopus H3K27 domains, which are relatively GC-poor. Interestingly, the frog-trained algorithm was also able to distinguish human and zebrafish H3K27me3 domains reasonably well. Similar training on human and zebrafish data revealed a large number of 8-mer motifs that are enriched or depleted in all three species, suggesting that despite the difference in GC content, vertebrate PREs may share common sequence principles.
Each of these predictive studies identified sequence features that are able to distinguish PcG targets from non-targets, and evaluated performance by comparing to ChIP enrichments (Arnold et al. 2013; Ku et al. 2008; van Heeringen et al. 2014). It would be interesting in future to evaluate sites predicted using these features that are not already covered by known ChIP enrichments, as these may lead to novel PREs that are not detected in the available ChIP datasets. In future, computational approaches such as these will be of high importance to identify the specific nucleation sites as opposed to those that are created by spreading or looping, to understand the minimal sequence requirements to establish a PcG domain, and to distinguish primary from secondary peaks (Fig. 6).
Conclusion and perspectives
In this review, we initially set out to address three questions, by comparing the protein and nucleic acid components of the PcG and TrxG regulatory system in flies and vertebrates: why are PREs not conserved? Do mammalian PREs use different sequences but perform essentially the same function as fly PREs? Or does mammalian PcG/TrxG regulation play by fundamentally different rules to those in the fly? In conclusion, we find that some of these questions can be answered better than others, and some in fact need to be reformulated. Figure 7 summarises the main findings of this analysis. In considering the properties of the PcG and TrxG protein complexes, remarkable similarities emerge. The most important conserved property is the potential for bistability: multiple self-reinforcing mechanisms for both the PcG and TrxG proteins exist, and opposite mechanisms antagonise each other, giving a potential molecular momentum to each extreme state, whilst destabilising intermediate states (Table 1). The conservation of these properties strongly suggests that the system can function similarly in flies and vertebrates. Whether it does so in specific contexts will depend on the quantitative contribution of forces that push the system towards one or other state.
Summary of similarities and differences relevant for fly and vertebrate PREs. The figure summarises the main points of this review. For PRC1, PRC2, TRXG, and target genes, key similarities and differences are listed, discussed in detail in the main text. For DNA-binding factors and PREs, open questions are identified, discussed in the conclusion section of the review
In answer to the question, “why are PREs not conserved?” we propose that this question needs to be reformulated to ask “do we know whether PREs are conserved?” Due to the large number of open questions regarding PRE sequence requirements in both flies and vertebrates, the answer to this second question is currently “no”. The major difference between fly and vertebrate genomes is the lack of DNA methylation in the former. In vertebrate genomes, CpG islands are oases of unmethylated DNA, allowing PcG and TrxG binding, but do these sites simply present a similar binding platform to vertebrate PcG and TrxG proteins as fly PREs do to the fly proteins? To answer this question, it would be extremely interesting to address the role of conserved ZF-CxxC proteins and of CpG dinucleotides at PREs in the fly. Clarification of the roles of conserved factors such as YY1/PHO, mGAF/GAF, REST/Charlatan, AEBP2 and RUNX1/Lozenge (Tables 2 and S1) will add illumination. Finally, there may be undiscovered targeting factors that bind sequences found in both fly and vertebrate PREs. For example, GT repeat stretches are highly enriched at both vertebrate and fly PcG-binding sites (Ringrose et al. 2003; Schuettengruber et al. 2009; van Heeringen et al. 2014) and these motifs have been shown to be essential for PRE-mediated silencing in the fly (Okulski et al. 2011). We ignore “uninteresting repeats” at our peril.
Finally the third question “do mammalian PREs use different sequences but perform essentially the same function as fly PREs?” cannot be answered on the basis of current evidence. Not only do we not understand the extent of similarity or difference in PRE sequence between flies and vertebrates, but the currently available assays for PRE function in vertebrates fall short of addressing fundamental PRE properties beyond PcG recruitment. In the future, it will be essential to devise assays to address whether and to what extent mammalian PREs are similar to fly PREs, namely, whether they are also TREs, whether they can indeed switch between states, and whether they mediate epigenetic memory. An understanding of the quantitative contribution of DNA sequence, genomic context and developmental signalling to these properties will require quantitative assays in which both recruitment and gene expression are monitored, using constructs integrated at identical genomic locations and which are evaluated at different stages of differentiation. Both the vertebrate and the fly fields may benefit from the use of emerging high-throughput assays (Akhtar et al. 2014; Krebs et al. 2014) to systematically dissect the relationship between PRE sequence, genomic context and function. We suspect that the “PRE code” may not be so different after all.
References
Akhtar W, Pindyurin AV, de Jong J, Pagie L, Ten Hoeve J, Berns A, Wessels LF, van Steensel B, van Lohuizen M (2014) Using TRIP for genome-wide position effect analysis in cultured cells. Nat Protoc 9(6):1255–1281
An S, Yeo KJ, Jeon YH, Song JJ (2011) Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism. J Biol Chem 286(10):8369–8374
Arnold P, Scholer A, Pachkov M, Balwierz PJ, Jorgensen H, Stadler MB, van Nimwegen E, Schubeler D (2013) Modeling of epigenome dynamics identifies transcription factors that mediate Polycomb targeting. Genome Res 23(1):60–73
Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML (2003) Transcription factor YY1 functions as a PcG protein in vivo. EMBO J 22(6):1347–1358
Basu A, Dasari V, Mishra RK, Khosla S (2014) The CpG island encompassing the promoter and first exon of human DNMT3L gene is a PcG/TrX response element (PRE). PLoS One 9(4):e93561
Bengani H, Ganapathi M, Singh GP, Brahmachari V (2007) Mining of putative cis-acting elements for chromatin mediated regulation of Hox genes in mammals by in-silico analysis. J Exp Zool B Mol Dev Evol 308(4):384–395
Bengani H, Mendiratta S, Maini J, Vasanthi D, Sultana H, Ghasemi M, Ahluwalia J, Ramachandran S, Mishra RK, Brahmachari V (2013) Identification and validation of a putative Polycomb responsive element in the human genome. PLoS One 8(6):e67217
Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006a) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125(2):315–326
Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD (2006b) Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol 26(7):2560–2569
Beuchle D, Struhl G, Muller J (2001) Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128(6):993–1004
Biggin MD, Bickel S, Benson M, Pirrotta V, Tjian R (1988) Zeste encodes a sequence-specific transcription factor that activates the ultrabithorax promoter in vitro. Cell 53(5):713–722
Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, Ishikura T, Long HK, Sheahan TW, Brockdorff N, Kessler BM, Koseki H, Klose RJ (2014) Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157(6):1445–1459
Blastyak A, Mishra RK, Karch F, Gyurkovics H (2006) Efficient and specific targeting of Polycomb group proteins requires cooperative interaction between grainyhead and pleiohomeotic. Mol Cell Biol 26(4):1434–1444
Boulard M, Edwards JR, Bestor TH (2015) FBXL10 protects Polycomb-bound genes from hypermethylation. Nat Genet 47(5):479–485
Bowers SR, Calero-Nieto FJ, Valeaux S, Fernandez-Fuentes N, Cockerill PN (2010) Runx1 binds as a dimeric complex to overlapping Runx1 sites within a palindromic element in the human GM-CSF enhancer. Nucleic Acids Res 38(18):6124–6134
Bowman SK, Deaton AM, Domingues H, Wang PI, Sadreyev RI, Kingston RE, Bender W (2014) H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex. Elife 3:e02833
Briggs MR, Kadonaga JT, Bell SP, Tjian R (1986) Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science 234(4772):47–52
Brinkman AB, Gu H, Bartels SJ, Zhang Y, Matarese F, Simmer F, Marks H, Bock C, Gnirke A, Meissner A, Stunnenberg HG (2012) Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res 22(6):1128–1138
Brockdorff N (2013) Noncoding RNA and Polycomb recruitment. RNA 19(4):429–442
Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, Kimura H, Ragoussis J, Teichmann SA, Pombo A (2012) Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell 10(2):157–170
Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA (1998) The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell 1(7):1057–1064
Brown JL, Fritsch C, Mueller J, Kassis JA (2003) The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development 130(2):285–294
Brown JL, Grau DJ, DeVido SK, Kassis JA (2005) An Sp1/KLF binding site is important for the activity of a Polycomb group response element from the Drosophila engrailed gene. Nucleic Acids Res 33(16):5181–5189
Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, Gabellini D (2012) A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell 149(4):819–831
Cao R, Zhang Y (2004) SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell 15(1):57–67
Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298(5595):1039–1043
Cao R, Tsukada Y, Zhang Y (2005) Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell 20(6):845–854
Caputo VS, Costa JR, Makarona K, Georgiou E, Layton DM, Roberts I, Karadimitris A (2013) Mechanism of Polycomb recruitment to CpG islands revealed by inherited disease-associated mutation. Hum Mol Genet 22(16):3187–3194
Cavalli G, Paro R (1998) The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 93(4):505–518
Cerase A, Smeets D, Tang YA, Gdula M, Kraus F, Spivakov M, Moindrot B, Leleu M, Tattermusch A, Demmerle J, Nesterova TB, Green C, Otte AP, Schermelleh L, Brockdorff N (2014) Spatial separation of Xist RNA and polycomb proteins revealed by superresolution microscopy. Proc Natl Acad Sci U S A 111(6):2235–2240
Chan CS, Rastelli L, Pirrotta V (1994) A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J 13(11):2553–2564
Cheutin T, Cavalli G (2014) Polycomb silencing: from linear chromatin domains to 3D chromosome folding. Curr Opin Genet Dev 25:30–37
Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT (2014) Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell 55(2):171–185
Cooper S, Dienstbier M, Hassan R, Schermelleh L, Sharif J, Blackledge NP, De Marco V, Elderkin S, Koseki H, Klose R, Heger A, Brockdorff N (2014) Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep 7(5):1456–1470
Creppe C, Palau A, Malinverni R, Valero V, Buschbeck M (2014) A CBX8-containing polycomb complex facilitates the transition to gene activation during ES cell differentiation. PLoS Genet 10(12):e1004851
Cuddapah S, Roh TY, Cui K, Jose CC, Fuller MT, Zhao K, Chen X (2012) A novel human polycomb binding site acts as a functional polycomb response element in Drosophila. PLoS One 7(5):e36365
Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111(2):185–196
Davidovich C, Zheng L, Goodrich KJ, Cech TR (2013) Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol 20(11):1250–1257
de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7(5):663–676
Deaton AM, Bird A (2011) CpG islands and the regulation of transcription. Genes Dev 25(10):1010–1022
Denissov S, Hofemeister H, Marks H, Kranz A, Ciotta G, Singh S, Anastassiadis K, Stunnenberg HG, Stewart AF (2014) Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development 141(3):526–537
Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, Ozato K, Sims RJ, Singer DS (2012) BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci U S A 109(18):6927–6932
Dietrich N, Lerdrup M, Landt E, Agrawal-Singh S, Bak M, Tommerup N, Rappsilber J, Sodersten E, Hansen K (2012) REST-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genet 8(3):e1002494
Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS (2008) Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18(9):1433–1445
Duboule D (2007) The rise and fall of Hox gene clusters. Development 134(14):2549–2560
Duboule D, Morata G (1994) Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet 10(10):358–364
Enderle D, Beisel C, Stadler MB, Gerstung M, Athri P, Paro R (2011) Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome Res 21(2):216–226
Endoh M, Endo TA, Endoh T, Isono K, Sharif J, Ohara O, Toyoda T, Ito T, Eskeland R, Bickmore WA, Vidal M, Bernstein BE, Koseki H (2012) Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet 8(7):e1002774
Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, Bickmore WA (2010) Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 38(3):452–464
Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, Koseki H, Brockdorff N, Ponting CP, Kessler BM, Klose RJ (2012) KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife 1:e00205
Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S (2003) Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17(15):1870–1881
Fitzgerald DP, Bender W (2001) Polycomb group repression reduces DNA accessibility. Mol Cell Biol 21(19):6585–6597
Francis NJ, Kingston RE, Woodcock CL (2004) Chromatin compaction by a polycomb group protein complex. Science 306(5701):1574–1577
Gambetta MC, Muller J (2014) O-GlcNAcylation prevents aggregation of the Polycomb group repressor polyhomeotic. Dev Cell 31(5):629–639
Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D (2012) PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45(3):344–356
Gil J, O’Loghlen A (2014) PRC1 complex diversity: where is it taking us? Trends Cell Biol 24(11):632–641
Golebiowski FM, Gorecki A, Bonarek P, Rapala-Kozik M, Kozik A, Dziedzicka-Wasylewska M (2012) An investigation of the affinities, specificity and kinetics involved in the interaction between the Yin Yang 1 transcription factor and DNA. FEBS J 279(17):3147–3158
Grau DJ, Chapman BA, Garlick JD, Borowsky M, Francis NJ, Kingston RE (2011) Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev 25(20):2210–2221
Gregory GD, Vakoc CR, Rozovskaia T, Zheng X, Patel S, Nakamura T, Canaani E, Blobel GA (2007) Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol 27(24):8466–8479
Grossniklaus U, Paro R (2014) Transcriptional silencing by polycomb-group proteins. Cold Spring Harb Perspect Biol 6(11):a019331
Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477(7364):295–300
Hagarman JA, Motley MP, Kristjansdottir K, Soloway PD (2013) Coordinate regulation of DNA methylation and H3K27me3 in mouse embryonic stem cells. PLoS One 8(1):e53880
Hajjari M, Behmanesh M, Jahani MM (2014) In silico finding of putative cis-acting elements for the tethering of Polycomb Repressive Complex2 in human genome. Bioinformation 10(4):187–190
Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K (2008) A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10(11):1291–1300
Harrison MM, Botchan MR, Cline TW (2010) Grainyhead and zelda compete for binding to the promoters of the earliest-expressed Drosophila genes. Dev Biol 345(2):248–255
Hauenschild A, Ringrose L, Altmutter C, Paro R, Rehmsmeier M (2008) Evolutionary plasticity of polycomb/trithorax response elements in Drosophila species. PLoS Biol 6(10):e261
He GP, Kim S, Ro HS (1999) Cloning and characterization of a novel zinc finger transcriptional repressor. A direct role of the zinc finger motif in repression. J Biol Chem 274(21):14678–14684
He J, Kallin EM, Tsukada Y, Zhang Y (2008) The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat Struct Mol Biol 15(11):1169–1175
He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y (2013) Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol 15(4):373–384
Hekimoglu B, Ringrose L (2009) Non-coding RNAs in polycomb/trithorax regulation. RNA Biol 6(2):129–137
Hekimoglu-Balkan B, Aszodi A, Heinen R, Jaritz M, Ringrose L (2012) Intergenic Polycomb target sites are dynamically marked by non-coding transcription during lineage commitment. RNA Biol 9(3):314–325
Herz HM, Mohan M, Garrett AS, Miller C, Casto D, Zhang Y, Seidel C, Haug JS, Florens L, Washburn MP, Yamaguchi M, Shiekhattar R, Shilatifard A (2012) Polycomb repressive complex 2-dependent and -independent functions of Jarid2 in transcriptional regulation in Drosophila. Mol Cell Biol 32(9):1683–1693
Herzog VA, Lempradl A, Trupke J, Okulski H, Altmutter C, Ruge F, Boidol B, Kubicek S, Schmauss G, Aumayr K, et al (2014) A strand-specific switch in noncoding transcription switches the function of a Polycomb/Trithorax response element. Nat Genet
Hodgson JW, Argiropoulos B, Brock HW (2001) Site-specific recognition of a 70-base-pair element containing d(GA)(n) repeats mediates bithoraxoid polycomb group response element-dependent silencing. Mol Cell Biol 21(14):4528–4543
Holler M, Westin G, Jiricny J, Schaffner W (1988) Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev 2(9):1127–1135
Hu D, Garruss AS, Gao X, Morgan MA, Cook M, Smith ER, Shilatifard A (2013) The Mll2 branch of the COMPASS family regulates bivalent promoters in mouse embryonic stem cells. Nat Struct Mol Biol 20(9):1093–1097
Huang DH, Chang YL (2004) Isolation and characterization of CHRASCH, a polycomb-containing silencing complex. Methods Enzymol 377:267–282
Huang DH, Chang YL, Yang CC, Pan IC, King B (2002) Pipsqueak encodes a factor essential for sequence-specific targeting of a polycomb group protein complex. Mol Cell Biol 22(17):6261–6271
Hunkapiller J, Shen Y, Diaz A, Cagney G, McCleary D, Ramalho-Santos M, Krogan N, Ren B, Song JS, Reiter JF (2012) Polycomb-like 3 promotes polycomb repressive complex 2 binding to CpG islands and embryonic stem cell self-renewal. PLoS Genet 8(3):e1002576
Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr AR, James KD, Turner DJ, Smith C, Harrison DJ, Andrews R, Bird AP (2010) Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet 6(9):e1001134
Jermann P, Hoerner L, Burger L, Schubeler D (2014) Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc Natl Acad Sci U S A 111(33):E3415–E3421
Jothi R, Cuddapah S, Barski A, Cui K, Zhao K (2008) Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res 36(16):5221–5231
Jung HR, Pasini D, Helin K, Jensen ON (2010) Quantitative mass spectrometry of histones H3.2 and H3.3 in Suz12-deficient mouse embryonic stem cells reveals distinct, dynamic post-translational modifications at Lys-27 and Lys-36. Mol Cell Proteomics 9(5):838–850
Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M, Muller J (2014) Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol 21(6):569–571
Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D (2010) Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev 24(23):2615–2620
Kaneko S, Son J, Bonasio R, Shen SS, Reinberg D (2014) Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev 28(18):1983–1988
Kassis JA (1994) Unusual properties of regulatory DNA from the Drosophila engrailed gene: three “pairing-sensitive” sites within a 1.6-kb region. Genetics 136(3):1025–1038
Kassis JA, Brown JL (2013) Polycomb group response elements in Drosophila and vertebrates. Adv Genet 81:83–118
Kellner WA, Van Bortle K, Li L, Ramos E, Takenaka N, Corces VG (2013) Distinct isoforms of the Drosophila Brd4 homologue are present at enhancers, promoters and insulator sites. Nucleic Acids Res 41(20):9274–9283
Kennison JA (2004) Introduction to Trx-G and Pc-G genes. Methods Enzymol 377:61–70
Kherrouche Z, Begue A, Stehelin D, Monte D (2001) Molecular cloning and characterization of the mouse E2F6 gene. Biochem Biophys Res Commun 288(1):22–33
Kim J, Kim J (2009) YY1’s longer DNA-binding motifs. Genomics 93(2):152–158
Kim CG, Swendeman SL, Barnhart KM, Sheffery M (1990) Promoter elements and erythroid cell nuclear factors that regulate alpha-globin gene transcription in vitro. Mol Cell Biol 10(11):5958–5966
Kim CA, Gingery M, Pilpa RM, Bowie JU (2002) The SAM domain of polyhomeotic forms a helical polymer. Nat Struct Biol 9(6):453–457
Kim CA, Sawaya MR, Cascio D, Kim W, Bowie JU (2005) Structural organization of a Sex-comb-on-midleg/polyhomeotic copolymer. J Biol Chem 280(30):27769–27775
Kim H, Kang K, Kim J (2009) AEBP2 as a potential targeting protein for polycomb repression complex PRC2. Nucleic Acids Res 37(9):2940–2950
Kingston RE, Tamkun JW (2014) Transcriptional regulation by trithorax-group proteins. Cold Spring Harb Perspect Biol 6(10):a019349
Klose RJ, Cooper S, Farcas AM, Blackledge NP, Brockdorff N (2013) Chromatin sampling–an emerging perspective on targeting polycomb repressor proteins. PLoS Genet 9(8):e1003717
Kozma G, Bender W, Sipos L (2008) Replacement of a Drosophila Polycomb response element core, and in situ analysis of its DNA motifs. Mol Gen Genomics 279(6):595–603
Krajewski WA, Nakamura T, Mazo A, Canaani E (2005) A motif within SET-domain proteins binds single-stranded nucleic acids and transcribed and supercoiled DNAs and can interfere with assembly of nucleosomes. Mol Cell Biol 25(5):1891–1899
Krebs AR, Dessus-Babus S, Burger L, Schubeler D (2014) High-throughput engineering of a mammalian genome reveals building principles of methylation states at CG rich regions. Elife 3:e04094
Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, Adli M, Kasif S, Ptaszek LM, Cowan CA, Lander ES, Koseki H, Bernstein BE (2008) Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4(10):e1000242
Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16(22):2893–2905
Kuzmichev A, Jenuwein T, Tempst P, Reinberg D (2004) Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell 14(2):183–193
Kyba M, Brock HW (1998) The SAM domain of polyhomeotic, RAE28, and scm mediates specific interactions through conserved residues. Dev Genet 22(1):74–84
Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, Verrijzer CP (2008) dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev 22(20):2799–2810
Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T (1997) Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J 16(11):3219–3232
Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y (2007) A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449(7163):689–694
Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, Brookes E, Pombo A, Fisher C, Skarnes WC, Snoek T, Bezstarosti K, Demmers J, Klose RJ, Casanova M, Tavares L, Brockdorff N, Merkenschlager M, Fisher AG (2010) Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol 12(6):618–624
Lanzuolo C, Orlando V (2012) Memories from the Polycomb group proteins. Annu Rev Genet 46:561–589
Lehmann L, Ferrari R, Vashisht AA, Wohlschlegel JA, Kurdistani SK, Carey M (2012) Polycomb repressive complex 1 (PRC1) disassembles RNA polymerase II preinitiation complexes. J Biol Chem 287(43):35784–35794
Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D (2010) Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 24(4):368–380
Lo SM, Ahuja NK, Francis NJ (2009) Polycomb group protein Suppressor 2 of Zeste is a functional homolog of posterior sex combs. Mol Cell Biol 29(2):515–525
Long HK, Blackledge NP, Klose RJ (2013a) ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochem Soc Trans 41(3):727–740
Long HK, Sims D, Heger A, Blackledge NP, Kutter C, Wright ML, Grützner F, Odom DT, Patient R, Ponting CP, Klose RJ (2013b) Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. Elife 2:e00348
Lynch MD, Smith AJ, De Gobbi M, Flenley M, Hughes JR, Vernimmen D, Ayyub H, Sharpe JA, Sloane-Stanley JA, Sutherland L, Meek S, Burdon T, Gibbons RJ, Garrick D, Higgs DR (2012) An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J 31(2):317–329
Mace KA, Pearson JC, McGinnis W (2005) An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science 308(5720):381–385
Maeda RK, Karch F (2006) The ABC of the BX-C: the bithorax complex explained. Development 133(8):1413–1422
Maeda RK, Karch F (2009) The bithorax complex of Drosophila an exceptional Hox cluster. Curr Top Dev Biol 88:1–33
Margueron R, Reinberg D (2011) The Polycomb complex PRC2 and its mark in life. Nature 469(7330):343–349
Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D (2008) Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 32(4):503–518
Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ (2009) Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461(7265):762–767
Matharu NK, Hussain T, Sankaranarayanan R, Mishra RK (2010) Vertebrate homologue of Drosophila GAGA factor. J Mol Biol 400(3):434–447
Mazzoni EO, Mahony S, Peljto M, Patel T, Thornton SR, McCuine S, Reeder C, Boyer LA, Young RA, Gifford DK, Wichterle H (2013) Saltatory remodeling of Hox chromatin in response to rostrocaudal patterning signals. Nat Neurosci 16(9):1191–1198
McCall K, Bender W (1996) Probes of chromatin accessibility in the Drosophila bithorax complex respond differently to Polycomb-mediated repression. EMBO J 15(3):569–580
Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE (2010) GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet 6(12):e1001244
Meng S, Luo M, Sun H, Yu X, Shen M, Zhang Q, Zhou R, Ju X, Tao W, Liu D, Deng H, Lu Z (2010) Identification and characterization of Bmi-1-responding element within the human p16 promoter. J Biol Chem 285(43):33219–33229
Mihaly J, Mishra RK, Karch F (1998) A conserved sequence motif in Polycomb-response elements. Mol Cell 1(7):1065–1066
Min J, Zhang Y, Xu RM (2003) Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 17(15):1823–1828
Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah SA, Di Croce L (2012) Nonoverlapping functions of the Polycomb group CBX family of proteins in embryonic stem cells. Cell Stem Cell 10(1):47–62
Mousavi K, Zare H, Wang AH, Sartorelli V (2012) Polycomb protein Ezh1 promotes RNA polymerase II elongation. Mol Cell 45(2):255–262
Muller J, Gaunt S, Lawrence PA (1995) Function of the Polycomb protein is conserved in mice and flies. Development 121(9):2847–2852
Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111(2):197–208
Niessen HE, Demmers JA, Voncken JW (2009) Talking to chromatin: post-translational modulation of polycomb group function. Epigenetics Chromatin 2(1):10
Noordermeer D, Duboule D (2013) Chromatin architectures and Hox gene collinearity. Curr Top Dev Biol 104:113–148
Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y (2002) A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296(5570):1132–1136
Okulski H, Druck B, Bhalerao S, Ringrose L (2011) Quantitative analysis of polycomb response elements (PREs) at identical genomic locations distinguishes contributions of PRE sequence and genomic environment. Epigenetics Chromatin 4:4
O’Loghlen A, Munoz-Cabello AM, Gaspar-Maia A, Wu HA, Banito A, Kunowska N, Racek T, Pemberton HN, Beolchi P, Lavial F, Masui O, Vermeulen M, Carroll T, Graumann J, Heard E, Dillon N, Azuara V, Snijders AP, Peters G, Bernstein E, Gil J (2012) MicroRNA regulation of CBX7 mediates a switch of Polycomb orthologs during ESC differentiation. Cell Stem Cell 10(1):33–46
Orlando V, Jane EP, Chinwalla V, Harte PJ, Paro R (1998) Binding of trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J 17(17):5141–5150
Ost A, Lempradl A, Casas E, Weigert M, Tiko T, Deniz M, Pantano L, Boenisch U, Itskov PM, Stoeckius M, Ruf M, Rajewsky N, Reuter G, Iovino N, Ribeiro C, Alenius M, Heyne S, Vavouri T, Pospisilik JA (2014) Paternal diet defines offspring chromatin state and intergenerational obesity. Cell 159(6):1352–1364
Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K (2010a) JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464(7286):306–310
Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, Skotte J, Wutz A, Porse B, Jensen ON, Helin K (2010b) Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res 38(15):4958–4969
Patsialou A, Wilsker D, Moran E (2005) DNA-binding properties of ARID family proteins. Nucleic Acids Res 33(1):66–80
Pedone PV, Ghirlando R, Clore GM, Gronenborn AM, Felsenfeld G, Omichinski JG (1996) The single Cys2-His2 zinc finger domain of the GAGA protein flanked by basic residues is sufficient for high-affinity specific DNA binding. Proc Natl Acad Sci U S A 93(7):2822–2826
Pemberton H, Anderton E, Patel H, Brookes S, Chandler H, Palermo R, Stock J, Rodriguez-Niedenführ M, Racek T, de Breed L, Stewart A, Matthews N, Peters G (2014) Genome-wide co-localization of Polycomb orthologs and their effects on gene expression in human fibroblasts. Genome Biol 15(2):R23
Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J (2009) Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139(7):1290–1302
Perez L, Barrio L, Cano D, Fiuza UM, Muzzopappa M, Milan M (2011) Enhancer-PRE communication contributes to the expansion of gene expression domains in proliferating primordia. Development 138(15):3125–3134
Petruk S, Sedkov Y, Johnston DM, Hodgson JW, Black KL, Kovermann SK, Beck S, Canaani E, Brock HW, Mazo A (2012) TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell 150(5):922–933
Pirrotta V, Li HB (2012) A view of nuclear Polycomb bodies. Curr Opin Genet Dev 22(2):101–109
Poux S, McCabe D, Pirrotta V (2001) Recruitment of Polycomb group chromatin complexes in Drosophila. Development 128(1):75–85
Reddington JP, Perricone SM, Nestor CE, Reichmann J, Youngson NA, Suzuki M, Reinhardt D, Dunican DS, Prendergast JG, Mjoseng H, Ramsahoye BH, Whitelaw E, Greally JM, Adams IR, Bickmore WA, Meehan RR (2013) Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol 14(3):R25
Ren X, Kerppola TK (2011) REST interacts with CBX proteins and regulates polycomb repressive complex 1 occupancy at RE1 elements. Mol Cell Biol 31(10):2100–2110
Richly H, Aloia L, Di Croce L (2011) Roles of the Polycomb group proteins in stem cells and cancer. Cell Death Dis 2:e204
Riising EM, Comet I, Leblanc B, Wu X, Johansen JV, Helin K (2014) Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell 55(3):347–360
Ringrose L (2007) Polycomb comes of age: genome-wide profiling of target sites. Curr Opin Cell Biol 19(3):290–297
Ringrose L, Paro R (2004) Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet 38(1):413–443
Ringrose L, Paro R (2007) Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134(2):223–232
Ringrose L, Rehmsmeier M, Dura JM, Paro R (2003) Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev Cell 5(5):759–771
Robinson AK, Leal BZ, Chadwell LV, Wang R, Ilangovan U, Kaur Y, Junco SE, Schirf V, Osmulski PA, Gaczynska M, Hinck AP, Demeler B, McEwen DG, Kim CA (2012) The growth-suppressive function of the polycomb group protein polyhomeotic is mediated by polymerization of its sterile alpha motif (SAM) domain. J Biol Chem 287(12):8702–8713
Rozovskaia T, Tillib S, Smith S, Sedkov Y, Rozenblatt-Rosen O, Petruk S, Yano T, Nakamura T, Ben-Simchon L, Gildea J, Croce CM, Shearn A, Canaani E, Mazo A (1999) Trithorax and ASH1 interact directly and associate with the trithorax group-responsive bxd region of the ultrabithorax promoter. Mol Cell Biol 19(9):6441–6447
Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M (2007) Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics 6(5):820–834
Sanulli S, Justin N, Teissandier A, Ancelin K, Portoso M, Caron M, Michaud A, Lombard B, da Rocha ST, Offer J, Loew D, Servant N, Wassef M, Burlina F, Gamblin SJ, Heard E, Margueron R (2015) Jarid2 methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol Cell 57(5):769–783
Sasai N, Kato Y, Kimura G, Takeuchi T, Yamaguchi M (2007) The Drosophila jumonji gene encodes a JmjC-containing nuclear protein that is required for metamorphosis. FEBS J 274(23):6139–6151
Scheuermann JC, Gutierrez L, Muller J (2012) Histone H2A monoubiquitination and Polycomb repression: the missing pieces of the puzzle. Fly 6(3):162–168
Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, Bunker RD, Wirth U, Bouwmeester T, Bauer A, Ly-Hartig N, Zhao K, Chan H, Gu J, Gut H, Fischle W, Muller J, Thoma NH (2011) Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42(3):330–341
Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A (2006) Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J 25(13):3110–3122
Schoenherr CJ, Anderson DJ (1995) The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267(5202):1360–1363
Schorderet P, Lonfat N, Darbellay F, Tschopp P, Gitto S, Soshnikova N, Duboule D (2013) A genetic approach to the recruitment of PRC2 at the HoxD locus. PLoS Genet 9(11):e1003951
Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G (2007) Genome regulation by polycomb and trithorax proteins. Cell 128(4):735–745
Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G (2009) Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7(1):e13
Schuettengruber B, Oded Elkayam N, Sexton T, Entrevan M, Stern S, Thomas A, Yaffe E, Parrinello H, Tanay A, Cavalli G (2014) Cooperativity, specificity, and evolutionary stability of polycomb targeting in Drosophila. Cell Rep 9(1):219–233
Sedaghat Y, Miranda WF, Sonnenfeld MJ (2002) The jing Zn-finger transcription factor is a mediator of cellular differentiation in the Drosophila CNS midline and trachea. Development 129(11):2591–2606
Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH (2008) EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32(4):491–502
Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH (2009) Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139(7):1303–1314
Simon JA, Kingston RE (2009) Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10(10):697–708
Simon JA, Kingston RE (2013) Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 49(5):808–824
Simon J, Chiang A, Bender W, Shimell MJ, O’Connor M (1993) Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev Biol 158(1):131–144
Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, Lipshitz HD, Cordes SP (2009) A vertebrate polycomb response element governs segmentation of the posterior hindbrain. Cell 138(5):885–897
Smigova J, Juda P, Krejci J, Raska I (2014) Structural basis of polycomb bodies. Folia Biol (Praha) 60(Suppl 1):13–20
Soshnikova N, Duboule D (2009a) Epigenetic regulation of vertebrate Hox genes: a dynamic equilibrium. Epigenetics 4(8):537–540
Soshnikova N, Duboule D (2009b) Epigenetic temporal control of mouse Hox genes in vivo. Science 324(5932):1320–1323
Squazzo SL, O’Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ (2006) Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res 16(7):890–900
Srinivasan L, Pan X, Atchison ML (2005) Transient requirements of YY1 expression for PcG transcriptional repression and phenotypic rescue. J Cell Biochem 96(4):689–699
Srivastava S, Puri D, Garapati HS, Dhawan J, Mishra RK (2013) Vertebrate GAGA factor associated insulator elements demarcate homeotic genes in the HOX clusters. Epigenetics Chromatin 6(1):8
Steffen PA, Ringrose L (2014) What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol 15(5):340–356
Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A (2007) Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9(12):1428–1435
Stojic L, Jasencakova Z, Prezioso C, Stutzer A, Bodega B, Pasini D, Klingberg R, Mozzetta C, Margueron R, Puri PL, Schwarzer D, Helin K, Fischle W, Orlando V (2011) Chromatin regulated interchange between polycomb repressive complex 2 (PRC2)-Ezh2 and PRC2-Ezh1 complexes controls myogenin activation in skeletal muscle cells. Epigenetics Chromatin 4:16
Stros M (2010) HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta 1799(1–2):101–113
Tanaka Y, Katagiri Z, Kawahashi K, Kioussis D, Kitajima S (2007) Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene 397(1–2):161–168
Tanay A, O’Donnell AH, Damelin M, Bestor TH (2007) Hyperconserved CpG domains underlie Polycomb-binding sites. Proc Natl Acad Sci U S A 104(13):5521–5526
Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, Turner DJ, Illingworth R, Bird A (2010) CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464(7291):1082–1086
Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ (2009) CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila polycomb silencing. Development 136(18):3131–3141
Tie F, Banerjee R, Conrad PA, Scacheri PC, Harte PJ (2012) Histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine 27. Mol Cell Biol 32(12):2323–2334
Tie F, Banerjee R, Saiakhova AR, Howard B, Monteith KE, Scacheri PC, Cosgrove MS, Harte PJ (2014) Trithorax monomethylates histone H3K4 and interacts directly with CBP to promote H3K27 acetylation and antagonize Polycomb silencing. Development 141(5):1129–1139
Trimarchi JM, Fairchild B, Wen J, Lees JA (2001) The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc Natl Acad Sci U S A 98(4):1519–1524
Trojer P, Cao AR, Gao Z, Li Y, Zhang J, Xu X, Li G, Losson R, Erdjument-Bromage H, Tempst P, Farnham PJ, Reinberg D (2011) L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol Cell 42(4):438–450
Tsuda L, Kaido M, Lim YM, Kato K, Aigaki T, Hayashi S (2006) An NRSF/REST-like repressor downstream of Ebi/SMRTER/Su(H) regulates eye development in Drosophila. EMBO J 25(13):3191–3202
Uv AE, Thompson CR, Bray SJ (1994) The Drosophila tissue-specific factor Grainyhead contains novel DNA-binding and dimerization domains which are conserved in the human protein CP2. Mol Cell Biol 14(6):4020–4031
van Heeringen SJ, Akkers RC, van Kruijsbergen I, Arif MA, Hanssen LL, Sharifi N, Veenstra GJ (2014) Principles of nucleation of H3K27 methylation during embryonic development. Genome Res 24(3):401–410
Vasanthi D, Nagabhushan A, Matharu NK, Mishra RK (2013) A functionally conserved Polycomb response element from mouse HoxD complex responds to heterochromatin factors. Sci Rep 3:3011
Wachter E, Quante T, Merusi C, Arczewska A, Stewart F, Webb S, Bird A (2014) Synthetic CpG islands reveal DNA sequence determinants of chromatin structure. Elife 3:e03397
Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, Torchia J, Krogan NJ, Reiter JF, Stanford WL (2010) Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell 6(2):153–166
Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y (2004a) Role of histone H2A ubiquitination in polycomb silencing. Nature 431(7010):873–878
Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS (2004b) Hierarchical recruitment of polycomb group silencing complexes. Mol Cell 14(5):637–646
Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472(7341):120–124
Wildonger J, Sosinsky A, Honig B, Mann RS (2005) Lozenge directly activates argos and klumpfuss to regulate programmed cell death. Genes Dev 19(9):1034–1039
Wilkinson FH, Park K, Atchison ML (2006) Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc Natl Acad Sci U S A 103(51):19296–19301
Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE (2010) A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 140(1):99–110
Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE (2013) Variable requirements for DNA-binding proteins at polycomb-dependent repressive regions in human HOX clusters. Mol Cell Biol 33(16):3274–3285
Wu X, Johansen JV, Helin K (2013) Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell 49(6):1134–1146
Xu J, Shao Z, Li D, Xie H, Kim W, Huang J, Taylor JE, Pinello L, Glass K, Jaffe JD, Yuan GC, Orkin SH (2015) Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol Cell 57(2):304–316
Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM (2010) Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38(5):662–674
Yu M, Mazor T, Huang H, Huang HT, Kathrein KL, Woo AJ, Chouinard CR, Labadorf A, Akie TE, Moran TB, Xie H, Zacharek S, Taniuchi I, Roeder RG, Kim CF, Zon LI, Fraenkel E, Cantor AB (2012) Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell 45(3):330–343
Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B (2011) H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 286(10):7983–7989
Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N, Li X, Xu M, Zhang Z, Niu T, Han Z, Chai J, Zhou XJ, Gao S, Zhu B (2012) Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science 337(6097):971–975
Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322(5902):750–756
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving humans or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
Fly and vertebrate DNA binding proteins: expanded version. This table is identical to Table 2 in the main review but contains additional detail on molecular function in column 3. DNA binding proteins that have been shown to play a role in PcG or TrxG regulation in flies (light green) and vertebrates (dark green) are listed. a) Proteins whose function in PcG or TrxG regulation was first defined in flies are listed. Each fly protein is followed by the vertebrate homolog, if known. b) Proteins whose function in PcG or TrxG regulation was first defined in vertebrate are listed. Each vertebrate protein is followed by the fly homolog, if known. Column 3: Molecular function. Evidence for a role in PcG or TrxG regulation is given if available. Column 4: Binding site. Consensus binding sites are listed, using the IUPAC code for non-conserved nucleotides http://www.bioinformatics.org/sms/iupac.html. R=A/G; Y=C/T; S=G/C; W=A/T; K=G/T; M=A/C; B=C/G/T; D=A/G/T; H=A/C/T; V=A/C/G; N=A/C/G/T. (DOCX 369 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Bauer, M., Trupke, J. & Ringrose, L. The quest for mammalian Polycomb response elements: are we there yet?. Chromosoma 125, 471–496 (2016). https://doi.org/10.1007/s00412-015-0539-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-015-0539-4