Abstract
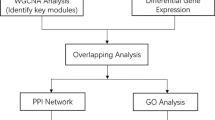
The aim of this study was to reveal a potential key gene network associated with seasonal allergic rhinitis (SAR). The microarray data GSE50101 downloaded from Gene Expression Omnibus were used to screen differentially expressed genes (DEGs) between SAR patients and healthy controls. Then, functional enrichment analysis was conducted using Database for Annotation, Visualization, and Integrated Discovery. Afterwards, the protein–protein interactions (PPIs) of DEGs were obtained from STRING, and the PPI network was constructed. In addition, the PPI network module was analyzed. In total, 98 up-regulated and 63 down-regulated DEGs were identified from the SAR samples, comparing the healthy controls. The up-regulated DEGs were mainly enriched in the Gene Ontology terms about cell death (e.g., DUSP1 and JUN) and pathways related to immune (e.g., FOS and JUN). The down-regulated DEGs were mainly enriched in regulation of transcription (e.g., CEBPD and SCML1). In the PPI network, a set of genes was predicted to interact with each other, such as FOS, JUN, and CEBPD. Furthermore, genes in the network module (e.g., FOS, JUN and CEBPD) was mainly enriched in regulation of transcription, and pathways about immune, such as mitogen-activated protein kinase signaling pathway, B cell receptor signaling pathway, and toll-like receptor signaling pathway. Several genes related to immunity and regulation of transcription, such as FOS, JUN, and CEBPD, may play crucial roles during the process of SAR through the interactions with each other.




Similar content being viewed by others
References
Leung KC, Hon KL (2013) Seasonal allergic rhinitis. Recent Pat Inflamm Allergy Drug Discov 7:187–201
Cavkaytar O, Akdis CA, Akdis M (2014) Modulation of immune responses by immunotherapy in allergic diseases. Curr Opin Pharmacol 17:30–37
Haenuki Y, Matsushita K, Futatsugi-Yumikura S, Ishii KJ, Kawagoe T, Imoto Y, Fujieda S, Yasuda M, Hisa Y, Akira S (2012) A critical role of IL-33 in experimental allergic rhinitis. J Allergy Clin Immunol 130(184–194):e111
Ando N, Nakamura Y, Ishimaru K, Ogawa H, Okumura K, Shimada S, Nakao A (2015) Allergen-specific basophil reactivity exhibits daily variations in seasonal allergic rhinitis. Allergy 70:319–322
Thornton MA, Akasheh N, Walsh M-T, Moloney M, Sheahan PO, Smyth CM, Walsh RM, Morgan RM, Curran DR, Walsh MT (2013) Eosinophil recruitment to nasal nerves after allergen challenge in allergic rhinitis. Clin Immunol 147:50–57
Nestor CE, Barrenas F, Wang H, Lentini A, Zhang H, Bruhn S, Jornsten R, Langston MA, Rogers G, Gustafsson M, Benson M (2014) DNA methylation changes separate allergic patients from healthy controls and may reflect altered CD4+ T-cell population structure. PLoS Genet 10:e1004059. doi:10.1371/journal.pgen.1004059
Smyth GK (2005) LIMMA: linear models for microarray data. In: Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, pp 397–420
Alvord G, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA (2007) The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8:R183
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452
Kohl M, Wiese S, Warscheid B (2011) Cytoscape: software for visualization and analysis of biological networks. In: Data mining in proteomics. Springer, New York, pp 291–303
Bader GD, Hogue CW (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform 4:2
Curran T, Franza BR (1988) Fos and Jun: the AP-1 connection. Cell 55:395–397
Angel P, Karin M (1991) The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta (BBA) Rev Cancer 1072:129–157
Salib R, Drake-Lee A, Howarth P (2003) Allergic rhinitis: past, present and the future. Clin Otolaryngol Allied Sci 28:291–303
Till S, Walker S, Dickason R, Huston D, O’brien F, Lamb J, Kay A, Corrigan C, Durham S (1997) IL-5 production by allergen-stimulated T cells following grass pollen immunotherapy for seasonal allergic rhinitis. Clin Exp Immunol 110:114–121
Ling EM, Smith T, Nguyen XD, Pridgeon C, Dallman M, Arbery J, Carr VA, Robinson DS (2004) Relation of CD4+ CD25+ regulatory T-cell suppression of allergen-driven T-cell activation to atopic status and expression of allergic disease. Lancet 363:608–615
Morse D, Pischke SE, Zhou Z, Davis RJ, Flavell RA, Loop T, Otterbein SL, Otterbein LE, Choi AM (2003) Suppression of inflammatory cytokine production by carbon monoxide involves the JNK pathway and AP-1. J Biol Chem 278:36993–36998
Ulanova M, Duta F, Puttagunta L, Schreiber AD, Befus AD (2005) Spleen tyrosine kinase (Syk) as a novel target for allergic asthma and rhinitis. Expert Opin Ther Targets 9:901–921
Jutel M, Blaser K, Akdis CA (2006) Histamine receptors in immune regulation and allergen-specific immunotherapy. Immunol Allergy Clin N Am 26:245–259
Barton GM, Medzhitov R (2003) Toll-like receptor signaling pathways. Science 300:1524–1525
Creticos PS, Schroeder JT, Hamilton RG, Balcer-Whaley SL, Khattignavong AP, Lindblad R, Li H, Coffman R, Seyfert V, Eiden JJ (2006) Immunotherapy with a ragweed—toll-like receptor 9 agonist vaccine for allergic rhinitis. N Engl J Med 355:1445–1455
Barrenäs F, Andersson B, Cardell LO, Langston M, Mobini R, Perkins A, Soini J, Ståhl A, Benson M (2008) Gender differences in inflammatory proteins and pathways in seasonal allergic rhinitis. Cytokine 42:325–329
Månsson A, Fransson M, Adner M, Benson M, Uddman R, Björnsson S, Cardell L-O (2010) TLR3 in human eosinophils: functional effects and decreased expression during allergic rhinitis. Int Arch Allergy Immunol 151:118–128
Luo X, Ma R, Wu X, Xian D, Li J, Mou Z, Li H (2015) Azelastine enhances the clinical efficacy of glucocorticoid by modulating MKP-1 expression in allergic rhinitis. Eur Arch Otorhinolaryngol 272:1165–1173
Balamurugan K, Sterneck E (2013) The many faces of C/EBP delta and their relevance for inflammation and cancer. Int J Biol Sci 9:917–933
Ko C-Y, Chang W-C, Wang J-M (2015) Biological roles of CCAAT/enhancer-binding protein delta during inflammation. J Biomed Sci 22:1–8
Zhang N, Truong-Tran QA, Tancowny B, Harris KE, Schleimer RP (2007) Glucocorticoids enhance or spare innate immunity: effects in airway epithelium are mediated by CCAAT/enhancer binding proteins. J Immunol 179:578–589
Johnson PF (2005) Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. J Cell Sci 118:2545–2555
Litvak V, Ramsey SA, Rust AG, Zak DE, Kennedy KA, Lampano AE, Nykter M, Shmulevich I, Aderem A (2009) Function of C/EBPδ in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol 10:437–443
Hofmann W-K, Tanosaki S, Gery S (2005) C/EBP delta expression in a BCR-ABL-positive cell line induces growth arrest and myeloid differentiation. Oncogene Int J 24:1589–1597
Scott L, Civin C, Rorth P, Friedman A (1992) A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood 80:1725–1735
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Funding
This work was supported by grants from China Postdoctoral Science Foundation funded project, by grants from the Priority Academic Program Development of Jiangsu Higher Education Institutions and by grants from Jiangsu Province Hospital of Traditional Chinese Medicine Program (Y16015).
Additional information
Jun Shi and Ying Zhang are co-first authors.
Rights and permissions
About this article
Cite this article
Shi, J., Zhang, Y., Qi, S. et al. Identification of potential crucial gene network related to seasonal allergic rhinitis using microarray data. Eur Arch Otorhinolaryngol 274, 231–237 (2017). https://doi.org/10.1007/s00405-016-4197-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00405-016-4197-9