Abstract
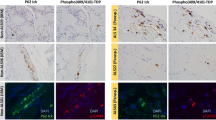
Intra-muscle fiber accumulation of ubiquitinated protein aggregates containing several conformationally modified proteins, including amyloid-β and phosphorylated tau, is characteristic of the pathologic phenotype of sporadic inclusion-body myositis (s-IBM), the most common progressive degenerative myopathy of older persons. Abnormalities of protein-degradation, involving both the 26S proteasome and autophagic-lysosomal pathways, were previously demonstrated in s-IBM muscle. NBR1 is a ubiquitin-binding scaffold protein importantly participating in autophagic degradation of ubiquitinated proteins. Whereas abnormalities of p62, a ubiquitin-binding protein, were previously described in s-IBM, abnormalities of NBR1 have not been reported in s-IBM. We have now identified in s-IBM muscle biopsies that NBR1, by: (a) immunohistochemistry, was strongly accumulated within s-IBM muscle-fiber aggregates, where it closely co-localized with p62, ubiquitin, and phosphorylated tau; (b) immunoblots, was increased threefold (p < 0.001); and (c) immunoprecipitation, was associated with p62 and LC3. By real-time PCR, NBR1 mRNA was increased twofold (p < 0.01). None of the various disease- and normal-control muscle biopsies had any NBR1 abnormality. In cultured human muscle fibers, NBR1 also physically associated with both p62 and LC3, and experimental inhibition of either the 26S proteasome or the lysosomal activity resulted in NBR1 increase. Our demonstration of NBR1 abnormalities in s-IBM provides further evidence that altered protein degradation pathways may be critically involved in the s-IBM pathogenesis. Accordingly, attempts to unblock defective protein degradation might be a therapeutic strategy for s-IBM patients.





Similar content being viewed by others
References
Askanas V, Alvarez RB, Engel WK (1993) Beta-amyloid precursor epitopes in muscle fibers of inclusion body myositis. Ann Neurol 34:551–560
Askanas V, Engel WK (1992) Cultured normal and genetically abnormal human muscle. In: Rowland LP, Di Mauro S (eds) The handbook of clinical neurology, myopathies, vol 18. North Holland, Amsterdam, pp 85–116
Askanas V, Engel WK (2008) Inclusion-body myositis: muscle-fiber molecular pathology and possible pathogenic significance of its similarity to Alzheimer’s and Parkinson’s disease brains. Acta Neuropathol 116:583–595
Askanas V, Engel WK (2011) Sporadic inclusion-body myositis: Conformational multifactorial ageing-related degenerative muscle disease associated with proteasomal and lysosomal inhibition, endoplasmic reticulum stress, and accumulation of amyloid-beta42 oligomers and phosphorylated tau. Presse Med 40:e219–e235
Askanas V, Engel WK, Alvarez RB, McFerrin J, Broccolini A (2000) Novel immunolocalization of alpha-synuclein in human muscle of inclusion-body myositis, regenerating and necrotic muscle fibers, and at neuromuscular junctions. J Neuropathol Exp Neurol 59:592–598
Askanas V, Engel WK, Nogalska A (2009) Inclusion body myositis: a degenerative muscle disease associated with intra-muscle fiber multi-protein aggregates, proteasome inhibition, endoplasmic reticulum stress and decreased lysosomal degradation. Brain Pathol 19:493–506
Bjorkoy G, Lamark T, Brech A et al (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171:603–614
Cecarini V, Bonfili L, Amici M, Angeletti M, Keller JN, Eleuteri AM (2008) Amyloid peptides in different assembly states and related effects on isolated and cellular proteasomes. Brain Res 1209:8–18
Ciechanover A (2011) Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Biochim Biophys Acta. doi:10.1016/j.bbapap.2011.03.007
Dalakas MC (2010) Inflammatory muscle diseases: a critical review on pathogenesis and therapies. Curr Opin Pharmacol 10:346–352
Ding WX, Yin XM (2008) Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy 4:141–150
Fratta P, Engel WK, McFerrin J, Davies KJA, Lin SW, Askanas V (2005) Proteasome inhibition and aggresome formation in sporadic inclusion-body myositis and in AbPP-overexpressing cultured human muscle fibers. Am J Pathol 167:517–526
Geisler S, Holmstrom KM, Skujat D et al (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12:119–131
He C, Klionsky DJ (2009) Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43:67–93
Iqbal K, Grundke-Iqbal I (2008) Alzheimer neurofibrillary degeneration: significance, etiopathogenesis, therapeutics and prevention. J Cell Mol Med 12:38–55
Itakura E, Mizushima N (2011) p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol 192:17–27
Johansen T, Lamark T (2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7:279–296
Keller JN, Hanni KB, Markesbery WR (2000) Impaired proteasome function in Alzheimer’s disease. J Neurochem 75:436–439
Kirkin V, Lamark T, Johansen T, Dikic I (2009) NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy 5:732–733
Kirkin V, Lamark T, Sou YS et al (2009) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 33:505–516
Klein WL, Stine WB Jr, Teplow DB (2004) Small assemblies of unmodified amyloid beta-protein are the proximate neurotoxin in Alzheimer’s disease. Neurobiol Aging 25:569–580
Klionsky DJ, Abeliovich H, Agostinis P et al (2008) Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4:151–175
Komatsu M, Waguri S, Koike M et al (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131:1149–1163
Kuusisto E, Salminen A, Alafuzoff I (2002) Early accumulation of p62 in neurofibrillary tangles in Alzheimer’s disease: possible role in tangle formation. Neuropathol App Neurobiol 28:228–237
LaFerla FM, Green KN, Oddo S (2007) Intracellular amyloid-beta in Alzheimer’s disease. Nat Rev Neurosci 8:499–509
Lamark T, Kirkin V, Dikic I, Johansen T (2009) NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle 8:1986–1990
Lange S, Xiang F, Yakovenko A et al (2005) The kinase domain of titin controls muscle gene expression and protein turnover. Science 308:1599–1603
Masiero E, Agatea L, Mammucari C et al (2009) Autophagy is required to maintain muscle mass. Cell Metab 10:507–515
Masiero E, Sandri M (2010) Autophagy inhibition induces atrophy and myopathy in adult skeletal muscles. Autophagy 6:307–309
Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM (1999) Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci USA 96:10403–10408
Mirabella M, Alvarez RB, Bilak M, Engel WK, Askanas V (1996) Difference in expression of phosphorylated tau epitopes between sporadic inclusion-body myositis and hereditary inclusion-body myopathies. J Neuropathol Exp Neurol 55:774–786
Mizushima N (2011) Autophagy in protein and organelle turnover. Cold Spring Harbor Symp Quant Biol. doi:10.1101/sqb.2011.76.011023
Nixon RA (2006) Autophagy in neurodegenerative disease: friend, foe or turncoat? Trends Neurosci 29:528–535
Nixon RA (2007) Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci 120:4081–4091
Nixon RA, Yang DS (2011) Autophagy failure in Alzheimer’s disease-locating the primary defect. Neurobiol Dis 43:38–45
Nogalska A, D’Agostino C, Engel WK, Askanas V (2011) Novel demonstration of conformationally-modified tau in sporadic inclusion-body myositis muscle fibers. Neurosci Lett. doi:10.1016/j.neulet.2011.08.042
Nogalska A, D’Agostino C, Engel WK, Davies KJ, Askanas V (2010) Decreased SIRT1 deacetylase activity in sporadic inclusion-body myositis muscle fibers. Neurobiol Aging 31:1637–1648
Nogalska A, D’Agostino C, Engel WK, Klein WL, Askanas V (2010) Novel demonstration of amyloid-beta oligomers in sporadic inclusion-body myositis muscle fibers. Acta Neuropathol 120:661–666
Nogalska A, D’Agostino C, Terracciano C, Engel WK, Askanas V (2010) Impaired autophagy in sporadic inclusion-body myositis and in endoplasmic reticulum stress-provoked cultured human muscle fibers. Am J Pathol 177:1377–1387
Nogalska A, Terracciano C, D’Agostino C, King Engel W, Askanas V (2009) p62/SQSTM1 is overexpressed and prominently accumulated in inclusions of sporadic inclusion-body myositis muscle fibers, and can help differentiating it from polymyositis and dermatomyositis. Acta Neuropathol 118:407–413
Pankiv S, Clausen TH, Lamark T et al (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282:24131–24145
Sandri M (2010) Autophagy in skeletal muscle. FEBS Lett 584:1411–1416
Sarkozi E, Askanas V, Engel WK (1994) Abnormal accumulation of prion protein mRNA in muscle fibers of patients with sporadic inclusion-body myositis and hereditary inclusion-body myopathy. Am J Pathol 145:1280–1284
Sarkozi E, Askanas V, Johnson SA, Engel WK, Alvarez RB (1993) beta-Amyloid precursor protein mRNA is increased in inclusion-body myositis muscle. Neuroreport 4:815–818
Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW (2004) Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol 24:8055–8068
Shacka JJ, Klocke BJ, Shibata M et al (2006) Bafilomycin A1 inhibits chloroquine-induced death of cerebellar granule neurons. Mol Pharmacol 69:1125–1136
Terracciano C, Nogalska A, Engel WK, Askanas V (2010) In AbetaPP-overexpressing cultured human muscle fibers proteasome inhibition enhances phosphorylation of AbetaPP751 and GSK3beta activation: effects mitigated by lithium and apparently relevant to sporadic inclusion-body myositis. J Neurochem 112:389–396
Tseng BP, Green KN, Chan JL, Blurton-Jones M, LaFerla FM (2008) A[beta] inhibits the proteasome and enhances amyloid and tau accumulation. Neurobiol Aging 29:1607–1618
Vattemi G, Nogalska A, King Engel W, D’Agostino C, Checler F, Askanas V (2009) Amyloid-beta42 is preferentially accumulated in muscle fibers of patients with sporadic inclusion-body myositis. Acta Neuropathol 117:569–574
Waters S, Marchbank K, Solomon E, Whitehouse C, Gautel M (2009) Interactions with LC3 and polyubiquitin chains link nbr1 to autophagic protein turnover. FEBS Lett 583:1846–1852
Weaver CL, Espinoza M, Kress Y, Davies P (2000) Conformational change as one of the earliest alterations of tau in Alzheimer’s disease. Neurobiol Aging 21:719–727
Wojcik S, Engel WK, Yan R, McFerrin J, Askanas V (2007) NOGO is increased and binds to BACE1 in sporadic inclusion-body myositis and in A beta PP-overexpressing cultured human muscle fibers. Acta Neuropathol 114:517–526
Zheng-Fischhèofer Q, Biernat J, Mandelkow EM, Illenberger S, Godemann R, Mandelkow E (1998) Sequential phosphorylation of Tau by glycogen synthase kinase-3beta and protein kinase A at Thr212 and Ser214 generates the Alzheimer-specific epitope of antibody AT100 and requires a paired-helical-filament-like conformation. Eur J Biochem 252:542–552
Acknowledgments
Maggie Baburyan provided technical assistance in electronmicroscopy. We are grateful to Dr Michael Jakowec of the USC Department of Neurology for allowing us to use his real-time PCR equipment. This work was supported by a grant from the Muscular Dystrophy Association to VA, and the Helen Lewis Research Fund.
Author information
Authors and Affiliations
Corresponding author
Additional information
Dr Nogalska is on leave from the Department of Biochemistry, Medical University of Gdansk, Gdansk, Poland. Mafalda Cacciottolo is a Ruth Ziegler Neuromuscular Fellow.
Rights and permissions
About this article
Cite this article
D’Agostino, C., Nogalska, A., Cacciottolo, M. et al. Abnormalities of NBR1, a novel autophagy-associated protein, in muscle fibers of sporadic inclusion-body myositis. Acta Neuropathol 122, 627–636 (2011). https://doi.org/10.1007/s00401-011-0874-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-011-0874-3