Abstract
This study assessed the effects of Reynoutria japonica, Rudbeckia laciniata, and Solidago gigantea invading sites within and outside river valleys on activity, biomass, and composition of soil microbial communities. Microbial properties such as soil respiration, urease and arylsulfatase activities, microbial biomass (based on substrate-induced respiration, or SIR, and phospholipid fatty acids, or PLFA), and community composition (based on PLFA) were determined. R. japonica encroached on sites characterized by the lowest values of microbiological properties and R. laciniata on sites with the highest microbiological quality. The effect of invasion on soil microbial properties depended on the invasive plant species. R. japonica significantly decreased microbial biomass, determined by both SIR and total PLFA, urease activity, fungal PLFA, fungal:bacterial PLFA ratio, gram-negative bacterial PLFA, and soil respiration in comparison to soil under adjacent native plant communities. Microbial community composition also differed between soils under R. japonica and those under native plants. In contrast, R. laciniata and S. gigantea did not influence most microbial properties, though S. gigantea significantly increased fungal PLFA and R. laciniata and S. gigantea increased fungal:bacterial PLFA ratio. The effects of plant invasion on microbial properties were basically similar in soils located within and outside river valleys, probably because initially (i.e., before invasion) soils from the two locations were largely similar in terms of basic properties such as texture, moisture, pH, C:N ratio, and most microbial properties.
Similar content being viewed by others
Introduction
Invasive plants alter ecosystem structure and functioning, as they differ from natives in the physiology, leaf-area allocation, shoot allocation, growth rate, size, and fitness (Ehrenfeld 2003; Liao et al. 2008; Van Kleunen et al. 2010). Plant invasions considerably reduce the diversity and abundance, or change the composition of aboveground and belowground communities, including soil microbial communities (Belnap and Phillips 2001; Yeates and Williams 2001; Broz et al. 2007; Kappes et al. 2007; Hejda et al. 2009; Moroń et al. 2009; Aguilera et al. 2010; Lenda et al. 2013). Indeed, invasive plants can change the composition or abundance of complex microbial communities and particular microbial groups, such as saprotrophic, mycorrhizal or pathogenic fungi, or ammonia-oxidizing bacteria (Kourtev et al. 2002; Hawkes et al. 2005; Batten et al. 2006; Broz et al. 2007; Niu et al. 2007; Zhang et al. 2009a, 2009b; Mincheva et al. 2014; Majewska et al. 2015). Crucial ecosystem processes, for example, organic matter decomposition and mineralization or nitrogen fixation, are also modified by invaders (Rice et al. 2004; Hawkes et al. 2005; Liao et al. 2008; Tharayil et al. 2013; Mincheva et al. 2014). These modifications may lead to changes in soil nutrient pools and element cycling, engendering positive feedback that potentially stabilizes current or accelerates further invasion, and prevent re-establishment of native species (Ehrenfeld 2003; Niu et al. 2007; Liao et al. 2008; Pii et al. 2015). The greater the invader-induced changes in soil, the greater the feedback that may be expected (Scharfy et al. 2010).
The magnitude and direction of changes in soil characteristics and processes brought about by invasion depend on local habitat conditions, invasive plant species traits, and/or the interaction of both (Meyerson et al. 2000; Ehrenfeld 2003; Koutika et al. 2007; Dassonville et al. 2008; Scharfy et al. 2009; Vilà et al. 2011; Hulme et al. 2013; Castro-Díez et al. 2014). Dassonville et al. (2008) reported that invasion increased nutrient concentrations of surface soils with initially low nutrient concentrations, while under the opposite conditions, a negative impact was mainly observed. Similarly, Scharfy et al. (2009) discovered that the effects of invasion on some soil physicochemical properties depended on P availability, being more pronounced at more P-rich sites.
Reynoutria japonica, Rudbeckia laciniata, and Solidago gigantea aggressively invade anthropogenic, semi-natural, and natural habitats in Europe, forming dense, near-monoculture stands. They exert negative influence on the biodiversity of valuable natural and extensively managed habitats (Zelnik 2012). These species are considered transformers (Tokarska-Guzik et al. 2010), i.e., invasive species that “change the character, condition, form, or nature of ecosystems over a substantial area” (Pyšek et al. 2004); therefore, their effects on soil microbial processes and thus soil functioning (Nannipieri et al. 2003) require detailed investigations. Previous research has been often conducted using either one or a small number of study sites, a single invasive species/genus, and/or measuring few microbial properties, making generalizations on the impact of the invasion on soil functioning problematic (Chapuis-Lardy et al. 2006; Herr et al. 2007; Scharfy et al. 2009, 2010; Aguilera et al. 2010; Dassonville et al. 2011; Tharayil et al. 2013; Mincheva et al. 2014). To the best of our knowledge, data on the influence of R. laciniata invasion on soil microbial properties are lacking.
We performed an extensive investigation based on 48 independent study sites to assess the effects of R. japonica, R. laciniata, and S. gigantea on the activity, biomass, and composition of soil microbial communities. The study sites were located either outside or within river valleys (on floodplains); the latter were affected by fluvial processes and expected to differ from the former in terms of microclimate, the presence of alluvial materials, and soil properties. We hypothesized that the invasion would influence soil microbial properties and thus soil functioning and that the direction and magnitude of the changes would depend on the invasive species identity and location (within or outside valleys).
Materials and methods
Invasive plant species
Three invasive plant species were selected for the study: R. japonica Houtt. [= Fallopia japonica (Houtt.) Ronse Decr., = Polygonum cuspidatum Sieb. & Zucc.] (Polygonaceae), R. laciniata L. (Asteraceae), and S. gigantea Aiton [= S. serotina Aiton] (Asteraceae). These perennial herbs can reach 1.2–3 m in height. R. japonica is native to eastern Asia, R. laciniata and S. gigantea to North America. They invade a range of habitats such as riparian zones around standing waters or streams, floodplain woods, forest clearings, swamps, wastelands, grasslands, cultivated field margins, fallows, roadsides, and/or areas along railroad tracks (Tokarska-Guzik et al. 2010; Sudnik-Wójcikowska 2011; Bzdęga et al. 2012; Zelnik 2012).
Study sites and soil sampling
Study sites were established on ca. 7500 km2 area in southern Poland (from 18° 55′ E to 20° 31′ E and from 49° 50′ N to 50° 22′ N; altitude from 180 to 320 m a.s.l.) (Fig. 1). The area lies in the transitional climate zone between a temperate oceanic climate in the west and a temperate continental climate in the east. Mean annual temperature fluctuates between 7 and 9 °C and precipitation between 700 and 900 mm. The vegetation season (days with average temperature >5 °C) spans a period between 210 and 230 days.
Forty-eight study sites were located within (N = 24) and outside (N = 24) river valleys (Fig. 1). The sites were localized relatively far from each other (a minimum of 0.4 km) and separated by areas overgrown by distinct plant communities in order to limit the possibility that the properties of one study site affect the properties of another. In other words, we did our best to have as independent study sites as possible (Fig. 1). Valley sites were localized in the floodplains of four main rivers, namely the Wisła, Raba, Skawa, and Skawinka, and their minor tributaries, at a maximum distance of ca. 10–50 m from the riverbed, depending on the size of the river. Therefore, sites within (in contrast to those outside) river valleys were potentially affected by fluvial processes. The two locations could be expected to differ in terms of microclimate and the presence of alluvial materials.
Of the forty-eight study sites, each site belonged to one of the three site types encroached by (1) R. japonica, Rj (N = 16), (2) R. laciniata, Rl (N = 16), or (3) S. gigantea, Sg (N = 16) (Fig. 1). All sites were dominated by tall herbaceous vegetation and characterized by compact and relatively large neighboring patches of invasive and native plants. At each study site, two paired plots (2 m × 2 m each) were established: one in a dense, near-monoculture (> 90 % cover) patch of the invasive plant and one in the adjacent native vegetation devoid of any invasive species (control). The contrasting (invaded vs. native) plots were located as close as possible, a maximum of 15 m from each other, so as to limit differences in soil properties between the two vegetation types existing prior to the invasion event, and at least 3 m from the edge of a patch so as to minimize any effects of contrasting vegetation such as shading or litterfall. Within each plot, vascular plants and bryophytes were recorded. The 12-grade cover-abundance scale was used (1, <0.2 % cover and 1 small individual; 2, <1 % and 1–3 small individuals; 3, 1–3 % and 2–5 individuals; 4, 3–5 % and 3–8 individuals; 5, 5–12 % and 8–20 individuals; 6, 13–25 %; 7, 25–35 %; 8, 36–50 %; 9, 51–65 %; 10, 66–75 %; 11, 76–85 %; 12, 86–100 %). Uninvaded vegetation did not differ significantly in terms of plant species number and total plant cover between valleys and outside-valley locations (Table 1). The uninvaded patches of vegetation in Rj and Sg valley sites were dominated by Phalaris arundinacea and Rubus caesius, followed by Petasites hybridus, Calystegia sepium, Aegopodium podagraria, and Urtica dioica (Rj valley sites) or Cirsium arvense and Tanacetum vulgare (Sg valley sites). Rl valley sites were dominated by Cirsium arvense, Galeopsis speciosa, Dactylis glomerata, and Chaerophyllum aromaticum. Calamagrostis epigejos, Arrhenatherum elatius, Cirsium arvense, and Equisetum arvense dominated both Rj and Sg outside-valley sites. These species were often accompanied by Rubus caesius (Rj outside-valley sites) or Agropyron repens and Tanacetum vulgare (Sg outside-valley sites). The patches of native vegetation in Rl outside-valley sites most often consisted of Arrhenatherum elatius, Cirsium arvense, Holcus lanatus and Centaurea phrygia.
In August 2013, three subsamples of topsoil (ca. 0–20 cm) were taken from each plot after the removal of organic layer consisting of fragmented litter and bulked to obtain one composite sample per plot. In total, 96 soil samples were collected: 3 site types (Rj vs. Rl vs. Sg) × 2 locations (within vs. outside valleys) × 8 replicates (sites) × 2 plots (invasive vs. native).
Laboratory work
Soil physicochemical properties
Soil samples were sieved (2 mm mesh) and divided into three parts that were then either dried for chemical analyses, frozen at −20 °C for phospholipid fatty acid (PLFA) analysis, or kept moist at 4 °C for the other microbial analyses. Dry weight was measured after drying overnight at 105 °C. Maximum water-holding capacity (WHC) was assessed using a gravimetric method. Soil texture was determined through a combination of sieving and sedimentation (ISO 11277 1998). Soil pH was measured in 1:5 (w:v) water suspensions (ISO 10390 1994) with a Hach Lange HQ40D meter. Organic C was determined with a Leco RC-612 and total S with a Leco SC-144 DR dry combustion analyzer. Total N was determined by Kjeldahl method; soil was digested in H2SO4 with Kjeltabs (K2SO4 + CuSO4·5H2O; Foss Tecator Digestor Auto) followed by distillation on a Foss Tecator Kjeltec 2300 Analyzer Unit. The available (Olsen) P was measured with an ion chromatograph (Dionex ICS-1100) following soil extraction with 0.5 M NaHCO3 (Olsen et al. 1954, modified).
Soil microbiological properties
The soil basal respiration rate and microbial biomass, determined by substrate-induced respiration (SIR), were measured in fresh soil samples adjusted to 50 % WHC. The samples were incubated in gastight jars at 22 °C for ca. 24 h; the evolved CO2 was absorbed in 0.2 M NaOH. The excess hydroxide was titrated with 0.1 M HCl following the addition of BaCl2 and phenolphthalein as an indicator. After the basal respiration measurements, a mixture of talcum and glucose monohydrate (4:1; 10 mg glucose g−1 dw soil) was added to the soil samples to measure substrate-induced respiration, which involves the determination of CO2 after 4 h of incubation (Beck et al. 1996, modified).
Urease activity was measured according to Kandeler (1996) using the unbuffered method. Soil samples were incubated at 37 °C for 2 h following the addition of urea solution. Released ammonium was extracted with KCl and determined by a modified Berthelot reaction, which is based on the reaction of sodium salicylate with ammonium in the presence of sodium dichloroisocyanurate and sodium nitroprusside at alkaline pH. Photometric measurements were performed at 690 nm with a Hach Lange DR 3800 spectrophotometer.
Arylsulfatase activity was measured according to Strobl and Traunmüller (1996). Following the addition of acetate buffer (pH = 5.8) and p-nitrophenylsulfate, soil samples were incubated at 37 °C for 1 h. Nitrophenol released by arylsulfatase activity was extracted, colored with NaOH, and determined at 420 nm using the Hach Lange DR 3800 spectrophotometer.
Phospholipid fatty acid (PLFA) analysis was performed according to Palojärvi (2006), with the exception of the lipid extraction, which was performed according to Macnaughton et al. (1997). Lipids were extracted from freeze-dried soil samples with a mixture of methanol:chloroform:phosphate buffer (2:1:0.8, v:v:v) using accelerated solvent extractor ASE 200 (Dionex; two 15-min cycles, 80 °C, 1200 PSI). Following the extraction, an appropriate volume of chloroform and deionized water was added to give the correct final ratio (chloroform:methanol:phosphate buffer/water; 1:1:0.9, v:v:v) and form two phases. The chloroform layer was transferred to a new test tube, evaporated under nitrogen at 37 °C, and stored at −20 °C. The lipids were separated into neutral-, glyco-, and phospholipids in Bakerbond silica gel SPE columns (500 mg, Baker) by eluting with chloroform, acetone, and methanol, respectively. The methanol fraction was reduced to dryness under nitrogen. The phospholipids were subjected to mild alkaline methanolysis, and the resulting fatty acid methyl esters were separated and identified using a Varian GC–MS system (Varian 3900 and Saturn 2100T) and NIST library. The CP-Select CB for FAME (50 m × 0.25 × 0.39) column (Agilent Technologies) was used. Helium was used as a carrier gas, and injections were made in split mode (1:100). Individual fatty acids were identified relative to several standards: 37-component FAME Mix (Supelco), Bacterial Acid Methyl Ester (BAME) Mix (Supelco), and a few additional one-component standards (Sigma-Aldrich). Methyl nonadecanoate (19:0; Fluka) was used as an internal standard.
Calculations and statistical analyses
C mic was calculated according to the equation: C mic (μg g−1) = 40.04x + 0.37, where x is the respiration rate given in microliters CO2 h−1 g−1 (Anderson and Domsch 1978).
Forty-eight PLFAs were measured in soil samples; however, most PLFAs were present in negligible amounts in soil. Eighteen fatty acids (14:0, 2OH 14:0, 15:0, a15:0, i15:0, 16:0, i16:0, 16:1ω7, 17:0, a17:0, i17:0, cy17:0, 17:1, 18:0, 18:1ω7, 18:1ω9, 18:1ω9t, 18:2ω6) with more than 0.5 % of the total relative abundance were present in most soils, and these fatty acids with cy19:0 were used in further calculations and statistical analyses (Bossio and Scow 1998; Moore-Kucera and Dick 2008). The sum of the above PLFAs (total PLFA) was used as a total microbial biomass indicator. PLFA 18:2ω6 was used as an indicator of saprophytic fungi. Bacteria were represented by 15:0, a15:0, i15:0, i16:0, 16:1ω7, 17:0, a17:0, i17:0, cy17:0, 18:1ω7, and cy19:0 (Frostegård and Bååth 1996; Ruess and Chamberlain 2010). The fatty acids a15:0, i15:0, i16:0, a17:0, and i17:0 were chosen to represent gram-positive and 16:1ω7, cy17:0, 18:1ω7, and cy19:0 gram-negative bacteria (Piotrowska-Seget and Mrozik 2003; Bardgett and Walker 2004; Moore-Kucera and Dick 2008; Cycoń et al. 2013).
Prior to statistical analyses, all variables were transformed by a logarithmic or exponential function and expressed on a 0–1 scale in order to achieve homogeneity of variances. Student’s t tests or Mann-Whitney U tests were used to compare uninvaded soil properties between valley and outside-valley sites. Principal component analysis (PCA) was performed on the concentrations of individual PLFAs, expressed in mole percentages, to arrive at the minimum number of variables, called principal components (PCs), representing the composition of microbial communities. One variable (PLFA 17:1) was excluded from the PCA due to its considerable deviation from a normal distribution. Interpretation of PCs was based on factor loadings of individual PLFAs on PCs. Split-plot ANOVA was used to examine the effect of site type (3 site types—Rj, Rl, and Sg, including both invaded and native plots; whole-plot factor), location (2 locations—within and outside valleys; whole-plot factor), and invasion (2 plot types within sites—invaded vs. native; split-plot factor), or their interactions, on soil microbiological properties. Split-plot ANOVA was followed by Tukey’s HSD test if any significant (p < 0.05) effects of site type, location, or their interactions on microbial indices were found. Contrast analysis was performed to test for differences in microbiological properties between invaded and native soils if any of the following interactions was significant: invasion × site type, invasion × location, or invasion × site type × location. Statistical analyses employed Statistica 9 (StatSoft Inc).
Results
Seven PCs with eigenvalues >1, explaining the highest percent of variance (a total of 77.8 % of variance in the PLFA data), were chosen to represent the microbial community composition. PC1 (19.9 % of variance) correlated mainly with i16:0 (−0.85), i17:0 (−0.74), 16:1ω7 (0.74), a17:0 (−0.54), cy17:0 (0.52), PC2 (15.1 %) with 16:0 (−0.84), cy17:0 (0.56), i15:0 (−0.54), a15:0 (0.53), a17:0 (0.53), PC3 (12.5 %) with 14:0 (−0.78), 15:0 (−0.76), 17:0 (−0.66), PC4 (10.3 %) with 18:2ω6 (0.64), a15:0 (−0.49), 18:1ω9t (0.45), PC5 (7.3 %) with 18:1ω9 (−0.49), 18:1ω7 (0.49), cy19:0 (0.49), PC6 (6.6 %) with 2OH 14:0 (−0.67), i17:0 (0.42), and PC7 (6.1 %) with 18:0 (0.59), cy19:0 (−0.50).
Most studied soils (70 %) were classified as clay, clay loam, or sandy clay loam. Uninvaded soils did not differ in terms of most basic physicochemical properties between valley and outside-valley sites (Table 1). Statistically significant (p < 0.05) differences between the locations were found only for Olsen P and C:S ratio. Uninvaded soils of the two locations were also similar in respect of nearly all microbiological properties, except urease activity (Table 1) and community composition represented by PC1 (p = 0.015).
Soil microbiological properties were mainly affected by two factors: site type and invasion × site type interaction (Table 2). Effects of other factors, i.e., location, invasion, site type × location, invasion × location, and invasion × site type × location, were either rarely significant or insignificant (Table 2).
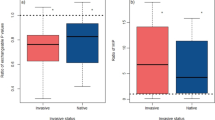
The effect of invasion on soil microbial communities depended strongly on the invasive plant species (significant effect of invasion × site type interaction for most microbiological properties; Table 2). R. japonica influenced many microbial indices. SIR-biomass, urease activity, total PLFA, fungal PLFA, fungal:bacterial PLFA ratio, gram-negative bacterial PLFA, and soil respiration were significantly lower under R. japonica than under adjacent native plant communities (Fig. 2). Microbial community composition also differed between soils from under R. japonica and native plants (p < 0.01 for PC2 and p < 0.001 for PC4 and PC7). R. laciniata and S. gigantea mainly influenced the abundance of fungal community, i.e., fungal PLFA and fungal:bacterial PLFA ratio (Fig. 2) and PC4 (p < 0.01 for S. gigantea), as well as PC6 (p < 0.05 for R. laciniata) and PC7 (p < 0.05 for R. laciniata and p < 0.01 for S. gigantea). More fungi were observed under R. laciniata and S. gigantea than under native communities, in contrast to the phenomenon observed for R. japonica.
Microbial properties of soils under invasive and native vegetation within and outside valleys (means and standard errors, N = 8). Different small letters above the bars denote statistically significant (p < 0.05) differences between three site types, i.e., sites encroached by R. japonica, R. laciniata, or S. gigantea, including both invaded and native plots. Asterisks indicate statistically significant effects of invasion, i.e., differences between invaded and native plots (***p < 0.001, **p < 0.01, *p < 0.05). The effect of invasion did not depend on location (within and outside valleys), with the exception of arylsulfatase activity. See Table 2 for results of the split-plot ANOVA
The influence of plant invasion on soil did not generally depend on location (Table 2). A significant effect of invasion × location interaction was only found in the case of PC1: microbial community composition was only influenced by invasion in outside-valley soils (p < 0.001). Moreover, arylsulfatase activity responded to invasion depending on both location and site type: it was significantly lower under R. japonica than under native plant communities in river valley soils (Table 2, Fig. 2).
The significant site type effect means that total microbial and bacterial biomass, enzyme activities, and community composition varied among sites of different invasive plant species, irrespectively of patch (plot) type, i.e., invaded vs. native; R. japonica encroached on sites characterized by the lowest SIR-biomass, enzyme activities, total PLFA, bacterial PLFA, and gram-positive bacterial PLFA, while the opposite was detected for R. laciniata (Fig. 2). Sites infested by R. japonica also differed from those encroached upon by R. laciniata in terms of the soil microbial community composition represented by PCs (p < 0.01 for PC1 and PC2). Most microbiological properties did not differ significantly between the locations. The only exceptions were urease activity, which was lower in outside-valley than in valley soils, and microbial community composition represented by PC1, PC4, and PC7 (Table 2).
Discussion
This study assessed the influence of three invasive plant species, R. japonica, R. laciniata, and S. gigantea, on activity, biomass, and composition of microbial communities in soils located within and outside river valleys. Our results clearly show that the invasion effects on soil microbes differed among the invasive plant species. R. japonica infestation significantly altered both the composition and the function of soil microbial communities, irrespective of location (valley or outside valley). Specifically, its invasion reduced soil respiration, both urease and arylsulfatase activities, the abundance of fungi and gram-negative bacteria, and the fungal:bacterial ratio. The negative effects of R. japonica invasion on soil microbial properties agree with those reported by others. Mincheva et al. (2014) discovered that R. japonica litter decomposed 3–4 times more slowly than the litter of native grassland species. Dassonville et al. (2011) and Tharayil et al. (2013) found that R. japonica, or other species of the Reynoutria genus, decreased enzyme activities related to N mineralization and/or denitrification, although the changes were season dependent (Tharayil et al. 2013). Our soils invaded by R. japonica, which is a nonmycorrhizal plant, showed a reduced abundance and species richness of arbuscular mycorrhizal fungi (AMF) (Zubek et al. unpublished). On the other hand, Aguilera et al. (2010) found no negative influence of R. japonica infestation on soil processes such as ammonification, nitrification, or total N mineralization.
Plants modify soil properties, including microbial processes, through deposition of litter and rhizodeposition that differ between species in terms of quantity and chemical quality (Chen et al. 2015). Invasive plants are often characterized by higher productivity than co-occurring native vegetation (Vilà et al. 2011; Jo et al. 2015). The biomass produced by R. japonica is 2–5 times greater and characterized by lower concentrations of essential nutrients such as N, P, K, Mg, or Mn than the biomass of native plants (Dassonville et al. 2008; Aguilera et al. 2010). Moreover, the tissues of R. japonica are characterized by relatively high C:N and lignin:N ratios at the time of senescence and litterfall (Aguilera et al. 2010; Mincheva et al. 2014). Hence, R. japonica provides the soil with huge amounts of low-quality litter (Aguilera et al. 2010; Mincheva et al. 2014), which is probably the primary reason for the severe decline in most soil microbial properties under the R. japonica monoculture discovered in this study. R. japonica invasion may have actually affected microbial properties through changes in soil chemical characteristics, as it altered the amount of soil N–NO3 ¯ in comparison to control soil (Stefanowicz et al. unpublished). The poor quality of litter produced by R. japonica and low microbial activity and decomposition rate accumulated large, undecomposed fragments of organic matter and increased the organic layer thickness under the plant (Maurel et al. 2010). The thickness of the organic layer reached ca. 20 cm at some of our study sites (data not shown).
The decomposition of detritus produced by R. japonica, characterized by high C:N and lignin:N ratios and relatively recalcitrant, is expected to be accomplished predominantly by soil fungi, not by bacteria (De Boer et al. 2005; Thorn and Lynch 2007). Surprisingly, our observations suggest that soil from under R. japonica was not beneficial to fungi, as soil fungal PLFA and fungal:bacterial ratio were considerably lower under R. japonica than under native plants. Previous research showed that the abundance, biomass, or composition of saprotrophic fungal communities found on decomposing litter in terrestrial and stream ecosystems differed between R. japonica and native plants, though it depended on study sites or decaying tissues (leaves vs. stems) (Lecerf et al. 2007; Mincheva et al. 2014). According to Mincheva et al. (2014), there were some striking differences in fungal communities: Phoma sp. was isolated in very high abundance from R. japonica litter, whereas it was absent in native litter. The opposite observation was made for Mucor sp.—one of the dominant taxa in native litter.
Modifications of the soil environment by invasions can depend not only on the chemical composition of plant litter but also on the release of secondary metabolites. R. japonica produces numerous compounds that inhibit the growth of pathogenic bacteria and fungi (Kim et al. 2005; Zhang et al. 2013). Phenolics released by R. japonica can also affect seasonal patterns of soil processes such as N mineralization (Tharayil et al. 2013). Thus, these substances may also explain the low microbial activity and biomass detected in soil from under R. japonica in our study.
The two other invasive species, i.e., Rudbeckia laciniata and Solidago gigantea, did not influence most soil microbial properties. They significantly affected only fungi-related properties, namely fungal PLFA (S. gigantea) or fungal:bacterial PLFA ratio (R. laciniata, S. gigantea). Both absolute and relative abundance of soil fungi increased due to the invasion, a phenomenon which is the opposite of that found in R. japonica. Both plant species increased mycorrhizal frequency but decreased species richness of AMF (Zubek et al. unpublished). Our results agree with those of Scharfy et al. (2009, 2010) and Quist et al. (2014) who found that S. gigantea invasion had no influence or decreased soil respiration, bacterial biomass, or phosphomonoesterase activity but increased fungal biomass and/or fungal:bacterial ratio. S. gigantea litter may support fungal rather than bacterial growth as it has lower tissue concentrations of most nutrients and higher C:N ratio than adjacent native vegetation (Vanderhoeven et al. 2006; Thorn and Lynch 2007; Dassonville et al. 2008). However, such differences in tissue nutrient concentrations were not found by other authors (Stefanowicz et al. unpublished). High fungal biomass detected in this study may suggest that S. gigantea stimulates fungal pathogens in its introduced range (Scharfy et al. 2010), as shown previously for other invasive species (Nijjer et al. 2007; Mangla et al. 2008). For example, Mangla et al. (2008) found that Chromolaena odorata, a destructive tropical invasive weed, accumulated high concentrations of the generalist soil-borne fungi, Fusarium semitectum, thereby creating a negative feedback to native plant species. However, another species of the Solidago genus, S. canadensis, suppressed fungal or fungi-like pathogens (Zhang et al. 2009b). Stimulation or inhibition of soil microorganisms, including fungal pathogens, may be driven through exudation of allelochemicals (Zhang et al. 2009b). The composition of secondary metabolites, which potentially affect activity and biomass of microorganisms, differs between plant species, including those belonging to the same genus (Kalemba and Thiem 2004). Essential oil extracted from S. gigantea contained 95 compounds, of which sesqiterpene (–)-cyclocolorenone was dominant; it had both antibacterial and antifungal properties (Jacyno et al. 1991; Kalemba et al. 2001; Kalemba and Thiem 2004). It is known, however, that some secondary metabolites exhibit antibacterial, but not antifungal, activity (Saeedi and Morteza-Semnani 2009), potentially altering fungi:bacteria ratio in soil.
Invasive plants may also affect activity and biomass of bacteria and fungi indirectly through changes in soil fauna that feed on microorganisms. It was proved that invasive plants can alter the abundance or species composition of soil and sediment invertebrates, including fungivorous or bacterivorous nematodes, collembola, enchytraeids, and mites (Talley and Levin 2001; Yeates and Williams 2001; Quist et al. 2014; Rusterholz et al. 2014). These changes would possibly influence microorganisms as, depending on species and grazing intensity and selectivity, soil invertabrates can inhibit or stimulate mycelial growth and alter species dominance and composition within microbial communities (Hedlund and Augustsson 1995; Crowther et al. 2011).
The lack of effects of S. gigantea or R. laciniata invasion on most soil microbial properties in our research was unexpected, as both species are considered, like R. japonica, “transformers” that modify ecosystems considerably (Pyšek et al. 2004; Tokarska-Guzik et al. 2010). Other studies on the influence of S. gigantea on soil microbial properties provided inconsistent results, with positive, negative, or no changes in soil properties due to the invasion. Chapuis-Lardy et al. (2006) and Koutika et al. (2007) found that S. gigantea enhanced soil respiration, C mineralization, and alkaline and acid phosphomonoesterase activities in comparison to native vegetation. Similar results were reported by Zhang et al. (2009a) for other species of the Solidago genus; S. canadensis invasion increased soil microbial biomass, basal and substrate-induced respiration, and functional diversity of microbial communities. On the other hand, Herr et al. (2007) pointed out that microbial biomass P and acid phosphomonoesterase activity did not change due to S. gigantea infestation, while alkaline phosphomonoesterase activity declined considerably.
Contrary to expectations, the effects of invasion on soil microbial properties did not differ between valley and outside-valley sites, probably because the two locations did not differ in terms of basic soil properties such as total C, N and S content, texture, pH, or most microbial properties. Presumably the influence of fluvial processes we had expected in the river floodplain was too weak to alter soil properties as well as plant species richness and cover considerably. Dassonville et al. (2008) and Scharfy et al. (2009) found that the direction and magnitude of changes in soil properties induced by invasion differed between study sites, but their sites showed differences in initial (pre-invasive) soil properties. Specifically, invasion increased nutrient concentrations of surface soils with initially low nutrient concentrations, while under the opposite conditions a negative impact was mainly observed (Dassonville et al. 2008). Scharfy et al. (2009) reported that some effects of S. gigantea on soil depended on soil P availability, being more pronounced at P-rich sites.
In this study, soil samples were collected from paired plots, which were established in the patches of an invader and native vegetation. Such a sampling scheme did not exclude the possibility of pre-invasion differences among invaded and uninvaded plots as already mentioned by Vanderhoeven et al. (2005), Carol Adair and Burke (2010), and Dostál et al. (2013). However, our experimental plan avoided these differences since paired plots were established within one site, as close as possible to each other, in flat terrain and far enough from shrubs or trees. Experiments involving sowing, or planting, invasive and native plants in uninvaded soil should be performed in addition to field studies. Such experiments, testing the effects of R. japonica, R. laciniata, and S. gigantea on soil properties, are in progress.
Conclusions
Investigations concerning the effects of invasive plants on soil microbial properties are often based on only a single invasive species and sampling schemes that are affected by pseudoreplication. In contrast, our study included three invasive plant species and two locations (valleys and outside valleys) and used replicate independent study sites scattered over a vast area. The invasion of R. japonica, R. laciniata, and S. gigantea influenced soil microbial properties compared to native plants, but the direction and magnitude of the changes depended on the invasive species. The effects of R. japonica were the most pronounced: this species reduced microbial activity and biomass. Such negative effects of plant invasions on soil are not a common phenomenon as invasive plants generally increase nutrient pools, microbial activity, and rates of soil processes such as litter decomposition, N mineralization, or nitrification (Liao et al. 2008; Vilà et al. 2011). Changes in soil properties brought about by plant invasions may induce positive or negative feedback with implications for acceleration or stabilization of these invasions, re-establishment of the native flora, and the functioning of whole ecosystems. The effects of the invasive species on soil properties did not differ between valley and outside-valley locations, probably because soils of the two locations had similar initial characteristics. Further research should study the impact of plant invasions on soil microbial properties also testing seasonal effects and including ecosystems with contrasting properties.
References
Aguilera AG, Alpert P, Dukes JS, Harrington R (2010) Impacts of the invasive plant Fallopia japonica (Houtt.) on plant communities and ecosystem processes. Biol Invasions 12:1243–1252
Anderson JPE, Domsch KH (1978) A physiological method for the quantitative measurement of microbial biomass in soils. Soil Biol Biochem 10:215–221
Bardgett RD, Walker LR (2004) Impact of coloniser plant species on the development of decomposer microbial communities following deglaciation. Soil Biol Biochem 36:555–559
Batten KM, Scow KM, Davies KF, Harrison SP (2006) Two invasive plants alter soil microbial community composition in serpentine grasslands. Biol Invasions 8:217–230
Beck T, Öhlinger R, Baumgarten A (1996) Substrate-induced respiration. In: Schinner F, Öhlinger R, Kandeler E, Margesin R (eds) Methods in soil biology. Springer-Verlag, Berlin Heidelberg, pp. 64–68
Belnap J, Phillips SL (2001) Soil biota in an ungrazed grassland: response to annual grass (Bromus tectorum) invasion. Ecol Appl 11:1261–1275
Bossio DA, Scow KM (1998) Impacts of carbon and flooding on soil microbial communities: phospholipid fatty acid profiles and substrate utilization patterns. Microb Ecol 35:265–278
Broz AK, Manter DK, Vivanco JM (2007) Soil fungal abundance and diversity: another victim of the invasive plant Centaurea maculosa. ISME J 1:763–765
Bzdęga K, Janiak A, Tarłowska S, Kurowska M, Tokarska-Guzik B, Szarejko I (2012) Unexpected genetic diversity of Fallopia japonica from Central Europe revealed after AFLP analysis. Flora 207:636–645
Carol Adair E, Burke IC (2010) Plant phenology and life span influence soil pool dynamics: Bromus tectorum invasion of perennial C3-C4 grass communities. Plant Soil 335:255–269
Castro-Díez P, Godoy O, Alonso A, Gallardo A, Saldaña A (2014) What explains variation in the impacts of exotic plant invasions on the nitrogen cycle? A meta-analysis. Ecol Lett 17:1–12
Chapuis-Lardy L, Vanderhoeven S, Dassonville N, Koutika LS, Meerts P (2006) Effect of the exotic invasive plant Solidago gigantea on soil phosphorus status. Biol Fertil Soils 42:481–489
Chen Y, Sun J, Xie F, Wang X, Cheng G, Lu X (2015) Litter chemical structure is more important than species richness in affecting soil carbon and nitrogen dynamics including gas emissions from an alpine soil. Biol Fertil Soils 51:791–800
Crowther TW, Boddy L, Jones TH (2011) Species-specific effects of soil fauna on fungal foraging and decomposition. Oecologia 167:535–545
Cycoń M, Wójcik M, Borymski S, Piotrowska-Seget Z (2013) Short-term effects of the herbicide napropamide on the activity and structure of the soil microbial community assessed by the multi-approach analysis. Appl Soil Ecol 66:8–18
Dassonville N, Vanderhoeven S, Vanparys V, Hayez M, Gruber W, Meerts P (2008) Impacts of alien invasive plants on soil nutrients are correlated with initial site conditions in NW Europe. Oecologia 157:131–140
Dassonville N, Guillaumaud N, Piola F, Meerts P, Poly F (2011) Niche construction by the invasive Asian knotweeds (species complex Fallopia): impact on activity, abundance and community structure of denitrifiers and nitrifiers. Biol Invasions 13:1115–1133
De Boer W, Folman LB, Summerbell RC, Boddy L (2005) Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev 29:795–811
Dostál P, Müllerová J, Pyšek P, Pergl J, Klinerová T (2013) The impact of an invasive plant changes over time. Ecol Lett 16:1277–1284
Ehrenfeld JG (2003) Effects of exotic plant invasions on soil nutrient cycling processes. Ecosystems 6:503–523
Frostegård A, Bååth E (1996) The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol Fertil Soils 22:59–65
Hawkes CV, Wren IF, Herman DJ, Firestone MK (2005) Plant invasion alters nitrogen cycling by modifying the soil nitrifying community. Ecol Lett 8:976–985
Hedlund K, Augustsson A (1995) Effects of enchytraeid grazing on fungal growth and respiration. Soil Biol Biochem 27:905–909
Hejda M, Pyšek P, Jarošík V (2009) Impact of invasive plants on the species richness, diversity and composition of invaded communities. J Ecol 97:393–403
Herr C, Chapuis-Lardy L, Dassonville N, Vanderhoeven S, Meerts P (2007) Seasonal effect of the exotic invasive plant Solidago gigantea on soil pH and P fractions. J Plant Nutr Soil Sci 170:729–738
Hulme PE, Pyšek P, Jarošík V, Pergl J, Schaffner U, Vilà M (2013) Bias and error in understanding plant invasion impacts. Trends Ecol Evol 28:212–218
ISO 10390 (1994) Soil quality—determination of pH. ISO, Geneve
ISO 11277 (1998) Soil quality—determination of particle size distribution in mineral soil material—method by sieving and sedimentation. ISO, Geneve
Jacyno JM, Montemurro N, Bates AD, Cutler HG (1991) Phytotoxic and antimicrobial properties of cyclocolorenone from Magnolia grandiflora L. J Agric Food Chem 39:1166–1168
Jo I, Fridley JD, Frank DA (2015) Linking above- and belowground resource use strategies for native and invasive species of temperate deciduous forests. Biol Invasions:1545–1554
Kalemba D, Thiem B (2004) Constituents of the essential oils of four micropropagated Solidago species. Flavour Fragr J 19:40–43
Kalemba D, Marschall H, Bradesi P (2001) Constituents of the essential oil of Solidago gigantea Ait. (giant goldenrod). Flavour Fragr J 16:19–26
Kandeler E (1996) Urease activity by colorimetric technique. In: Schinner F, Öhlinger R, Kandeler E, Margesin R (eds) Methods in soil biology. Springer-Verlag, Berlin, pp. 171–174
Kappes H, Lay R, Topp W (2007) Changes in different trophic levels of litter-dwelling macrofauna associated with giant knotweed invasion. Ecosystems 10:734–744
Kim YS, Hwang CS, Shin DH (2005) Volatile constituents from the leaves of Polygonum cuspidatum S. et Z. and their anti-bacterial activities. Food Microbiol 22:139–144
Kourtev PS, Ehrenfeld JG, Huang WZ (2002) Enzyme activities during litter decomposition of two exotic and two native plant species in hardwood forests of New Jersey. Soil Biol Biochem 34:1207–1218
Koutika LS, Vanderhoeven S, Chapuis-Lardy L, Dassonville N, Meerts P (2007) Assessment of changes in soil organic matter after invasion by exotic plant species. Biol Fertil Soils 44:331–341
Lecerf A, Patfield D, Boiché A, Riipinen MP, Chauvet E, Dobson M (2007) Stream ecosystems respond to riparian invasion by Japanese knotweed (Fallopia japonica). Can J Fish Aquat Sci 64:1273–1283
Lenda M, Witek M, Skórka P, Moroń D, Woyciechowski M (2013) Invasive alien plants affect grassland ant communities, colony size and foraging behaviour. Biol Invasions 15:2403–2414
Liao C, Peng R, Luo Y, Zhou X, Wu X, Fang C, Chen J, Li B (2008) Altered ecosystem carbon and nitrogen cycles by plant invasion: a meta-analysis. New Phytol 177:706–714
Macnaughton SJ, Jenkins TL, Wimpee MH, Cormiér MR, White DC (1997) Rapid extraction of lipid biomarkers from pure culture and environmental samples using pressurized hot solvent extraction. J Microbiol Methods 31:19–27
Majewska ML, Błaszkowski J, Nobis M, Rola K, Nobis A, Łakomiec D, Czachura P, Zubek S (2015) Root-inhabiting fungi in alien plant species in relation to invasion status and soil chemical properties. Symbiosis 65:101–115
Mangla S, Inderjit, Callaway RM (2008) Exotic invasive plant accumulates native soil pathogens which inhibit native plants. J Ecol 96:58–67
Maurel N, Salmon S, Ponge JF, Machon N, Moret J, Muratet A (2010) Does the invasive species Reynoutria japonica have an impact on soil and flora in urban wastelands? Biol Invasions 12:1709–1719
Meyerson LA, Saltonstall K, Windham L, Kiviat E, Findlay S (2000) A comparison of Phragmites australis in freshwater and brackish marsh environments in North America. Wetl Ecol Manag 8:89–103
Mincheva T, Barni E, Varese GC, Brusa G, Cerabolini B, Siniscalco C (2014) Litter quality, decomposition rates and saprotrophic mycoflora in Fallopia japonica (Houtt.) Ronse Decraene and in adjacent native grassland vegetation. Acta Oecol 54:29–35
Moore-Kucera J, Dick RP (2008) PLFA profiling of microbial community structure and seasonal shifts in soils of a Douglas-fir chronosequence. Microb Ecol 55:500–511
Moroń D, Lenda M, Skórka P, Szentgyörgyi SJ, Woyciechowski M (2009) Wild pollinator communities are negatively affected by invasion of alien goldenrods in grassland landscapes. Biol Conserv 142:1322–1332
Nannipieri P, Ascher J, Ceccherini MT, Landi L, Pietramellara G, Renella G (2003) Microbial diversity and soil functions. Eur J Soil Sci 54:655–670
Nijjer S, Rogers WE, Siemann E (2007) Negative plant-soil feedbacks may limit persistence of an invasive tree due to rapid accumulation of soil pathogens. Proc R Soc B 274:2621–2627
Niu HB, Liu WX, Wan FH, Liu B (2007) An invasive aster (Ageratina adenophora) invades and dominates forest understories in China: altered soil microbial communities facilitate the invader and inhibit natives. Plant Soil 294:73–85
Olsen SR, Cole CV, Watanabe FS, Dean LA (1954) Estimation of available phosphorus in soils by extraction with sodium bicarbonate. Circular No 939, United States Department of Agriculture, Washington
Palojärvi A (2006) Phospholipid fatty acid (PLFA) analyses. In: Bloem J, Hopkins DW, Benedetti A (eds) Microbiological methods for assessing soil quality. Cab International, Wallingford, UK, pp. 204–211
Pii Y, Mimmo T, Tomasi N, Terzano R, Cesco S, Crecchio C (2015) Microbial interactions in the rhizosphere: beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol Fertil Soils 51:403–415
Piotrowska-Seget Z, Mrozik A (2003) Signature lipid biomarker (SLB) analysis in determining changes in community structure of soil microorganisms. Pol J Environ Stud 12:669–675
Pyšek P, Richardson DM, Rejmánek M, Webster GL, Williamson M, Kirschner J (2004) Alien plants in checklists and floras: towards better communication between taxonomists and ecologists. Taxon 53:131–143
Quist CW, Vervoort MTW, Van Megen H, Gort G, Bakker J, Van der Putten WH, Helder J (2014) Selective alteration of soil food web components by invasive giant goldenrod Solidago gigantea in two distinct habitat types. Oikos 123:837–845
Rice SK, Westerman B, Federici R (2004) Impacts of the exotic, nitrogen-fixing black locust (Robinia pseudoacacia) on nitrogen-cycling in a pine-oak ecosystem. Plant Ecol 174:97–107
Ruess L, Chamberlain PM (2010) The fat that matters: soil food web analysis using fatty acids and their carbon stable isotope signature. Soil Biol Biochem 42:1898–1910
Rusterholz HP, Salamon JA, Ruckli R, Baur B (2014) Effects of the annual invasive plant Impatiens glandulifera on the Collembola and Acari communities in a deciduous forest. Pedobiologia 57:285–291
Saeedi M, Morteza-Semnani K (2009) Chemical composition and antimicrobial activity of essential oil of Hyssopus angustifolius M.B. J Essent Oil Bear Plants 12:111–119
Scharfy D, Eggenschwiler H, Olde Venterink H, Edwards PJ, Güsewell (2009) The invasive alien plant species Solidago gigantea alters ecosystem properties across habitats with differing fertility. J Veg Sci 20:1072–1085
Scharfy D, Güsewell S, Gessner MO, Olde Venterink H (2010) Invasion of Solidago gigantea in contrasting experimental plant communities: effects on soil microbes, nutrients and plant-soil feedbacks. J Ecol 98:1379–1388
Strobl W, Traunmüller M (1996) Arylsulfatase activity. In: Schinner F, Öhlinger R, Kandeler E, Margesin R (eds) Methods in soil biology. Springer-Verlag, Berlin, pp. 230–232
Sudnik-Wójcikowska B (2011) Rośliny synantropijne. Multico Oficyna Wydawnicza, Warszawa
Talley TS, Levin LA (2001) Modification of sediments and macrofauna by an invasive marsh plant. Biol Invasions 3:51–68
Tharayil N, Alpert P, Bhowmik P, Gerard P (2013) Phenolic inputs by invasive species could impart seasonal variations in nitrogen pools in the introduced soils: a case study with Polygonum cuspidatum. Soil Biol Biochem 57:858–867
Thorn RG, Lynch MDJ (2007) Fungi and eukaryotic algae. In: Paul EA (ed) Soil microbiology and biochemistry. Elsevier, Amsterdam Boston Heidelberg London New York Oxford Paris San Diego San Francisco Singapore Sydney Tokyo, pp. 145–162
Tokarska-Guzik B, Węgrzynek B, Urbisz A, Urbisz A, Nowak T, Bzdęga K (2010) Alien vascular plants in the Silesian Upland of Poland: distribution, patterns, impacts and threats. Biodiv Res Conserv 19:33–54
Van Kleunen M, Weber E, Fischer M (2010) A meta-analysis of trait differences between invasive and non-invasive plant species. Ecol Lett 13:235–245
Vanderhoeven S, Dassonville N, Meerts P (2005) Increased topsoil mineral nutrient concentrations under exotic invasive plants in Belgium. Plant Soil 275:169–179
Vanderhoeven S, Dassonville N, Chapuis-Lardy L, Hayez M, Meerts P (2006) Impact of the invasive alien plant Solidago gigantea on primary productivity, plant nutrient content and soil mineral nutrient concentrations. Plant Soil 286:259–268
Vilà M, Espinar JL, Hejda M, Hulme PE, Jarošik V, Maron JL, Pergl J, Schaffner U, Sun Y, Pyšek P (2011) Ecological impacts of invasive alien plants: a meta-analysis of their effects on species, communities and ecosystems. Ecol Lett 14:702–708
Yeates GW, Williams PA (2001) Influence of three invasive weeds and site factors on soil microfauna in New Zealand. Pedobiologia 45:367–383
Zelnik I (2012) The presence of invasive alien plant species in different habitats: case study from Slovenia. Acta Biol Slov 55:25–38
Zhang CB, Wang J, Qian BY, Li WH (2009a) Effects of the invader Solidago canadensis on soil properties. Appl Soil Ecol 43:163–169
Zhang S, Jin Y, Tang J, Chen X (2009b) The invasive plant Solidago canadensis L. suppresses local soil pathogens through allelopathy. Appl Soil Ecol 41:215–222
Zhang L, Ravipati AS, Koyyalamudi SR, Jeong SC, Reddy N, Bartlett J, Smith PT, de la Cruz M, Monteiro MC, Melguizo Á, Jiménez E, Vicente F (2013) Anti-fungal and anti-bacterial activities of ethanol extracts of selected traditional Chinese medicinal herbs. Asian Pac J Trop Med 6:673–681
Acknowledgments
The authors would like to thank Elżbieta Chrzanowska, Stanisław Braniewski, and Urszula Korzeniak for their valuable help with the laboratory work. We thank Sławomir Borymski and Maciej Choczyński for the advice on PLFA analysis and Irena M. Grześ for her critical reading of an earlier version of this manuscript. The research received financial support from the National Science Center of the Republic of Poland, under the project DEC–2011/03/B/NZ8/00008 (2012–16). The W. Szafer Institute of Botany, Polish Academy of Sciences, also provided partial funding.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Stefanowicz, A.M., Stanek, M., Nobis, M. et al. Species-specific effects of plant invasions on activity, biomass, and composition of soil microbial communities. Biol Fertil Soils 52, 841–852 (2016). https://doi.org/10.1007/s00374-016-1122-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-016-1122-8