Abstract
Key message
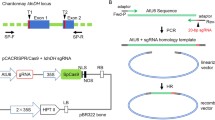
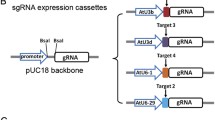
CRISPR/Cas9 system can precisely edit genomic sequence and effectively create knockout mutations in T0 generation watermelon plants.
Abstract
Genome editing offers great advantage to reveal gene function and generate agronomically important mutations to crops. Recently, RNA-guided genome editing system using the type II clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (Cas9) has been applied to several plant species, achieving successful targeted mutagenesis. Here, we report the genome of watermelon, an important fruit crop, can also be precisely edited by CRISPR/Cas9 system. ClPDS, phytoene desaturase in watermelon, was selected as the target gene because its mutant bears evident albino phenotype. CRISPR/Cas9 system performed genome editing, such as insertions or deletions at the expected position, in transfected watermelon protoplast cells. More importantly, all transgenic watermelon plants harbored ClPDS mutations and showed clear or mosaic albino phenotype, indicating that CRISPR/Cas9 system has technically 100% of genome editing efficiency in transgenic watermelon lines. Furthermore, there were very likely no off-target mutations, indicated by examining regions that were highly homologous to sgRNA sequences. Our results show that CRISPR/Cas9 system is a powerful tool to effectively create knockout mutations in watermelon.



Similar content being viewed by others
References
Brooks C, Nekrasov V, Lippman ZB, van Eck J (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol 166:1292–1297
Cardi T, Stewart CN Jr (2016) Progress of targeted genome modification approaches in higher plants. Plant Cell Rep 35:1401–1416
Chandrasekaran J, Brumin M, Wolf D, Leibman D, Klap C, Pearlsman M, Sherman A, Arazi T, Gal-On A (2016) Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol 17:1140–1153
Cho MA, Moon CY, Liu JR, Choi PS (2008) Agrobacterium-mediated transformation in Citrullus lanatus. Biol Plant 52:365–369
Choi PS, Soh WY, Kim YS, Yoo OJ, Liu JR (1994) Genetic transformation and plant regeneration of watermelon using Agrobacterium tumefaciens. Plant Cell Rep 13:344–348
Ellul P, Rios G, Atares A, Roig LA, Serrano R, Moreno V (2003) The expression of the Saccharomyces cerevisiae HAL1 gene increases salt tolerance in transgenic watermelon [Citrullus lanatus (Thunb.) Matsun. & Nakai.]. Theor Appl Genet 107:462–469
Fan D, Liu T, Li C, Jiao B, Li S, Hou Y, Luo K (2015) Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci Rep 5:12217
Gao J, Wang G, Ma S, Xie X, Wu X, Zhang X, Wu Y, Zhao P, Xia Q (2015) CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol Biol 87:99–110
Guo S, Zhang J, Sun H, Salse J, Lucas WJ, Zhang H, Zheng Y, Mao L, Ren Y, Wang Z, Min J, Guo X, Murat F, Ham BK, Zhang Z, Gao S, Huang M, Xu Y, Zhong S, Bombarely A, Mueller LA, Zhao H, He H, Zhang Y, Zhang Z, Huang S, Tan T, Pang E, Lin K, Hu Q, Kuang H, Ni P, Wang B, Liu J, Kou Q, Hou W, Zou X, Jiang J, Gong G, Klee K, Schoof H, Huang Y, Hu X, Dong S, Liang D, Wang J, Wu K, Xia Y, Zhao X, Zheng Z, Xing M, Liang X, Huang B, Lv T, Wang J, Yin Y, Yi H, Li R, Wu M, Levi A, Zhang X, Giovannoni JJ, Wang J, Li Y, Fei Z, Xu Y (2013) The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet 45:51–58
Huang YC, Chiang CH, Li CM, Yu TA (2011) Transgenic watermelon lines expressing the nucleocapsid gene of Watermelon silver mottle virus and the role of thiamine in reducing hyperhydricity in regenerated shoots. Plant Cell Tissue Organ Cult 106:21–29
Jia Y, Ding Y, Shi Y, Zhang X, Gong Z, Yang S (2016) The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol 212:345–353
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Lee J, Chung JH, Kim HM, Kim DW, Kim H (2016) Designed nucleases for targeted genome editing. Plant Biotechnol J 14:448–462
Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31:688–691
Li J, Meng X, Zong Y, Chen K, Zhang H, Liu J, Li J, Gao C (2016) Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat Plants 2:16139
Liu L, Gu Q, Ijaz R, Zhang J, Ye Z (2016) Generation of transgenic watermelon resistance to Cucumber mosaic virus facilitated by an effective Agrobacterium -mediated transformation method. Sci Hortic 205:32–38
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8:1274–1284
Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ (2015) CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods 12:982–988
Nishibayashi S, Kaneko H, Hayakawa T (1996) Transformation of cucumber (Cucumis sativus L.) plants using Agrobacterium tumefaciens and regeneration from hypocotyl explants. Plant Cell Rep 15:809–814
Qin G, Gu H, Ma L, Peng Y, Deng XW, Chen Z, Qu LJ (2007) Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res 17:471–482
Ren C, Liu X, Zhang Z, Wang Y, Duan W, Li S, Liang Z (2016) CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.). Sci Rep 6:32289
Schaeffer SM, Nakata PA (2016) The expanding footprint of CRISPR/Cas9 in the plant sciences. Plant Cell Rep 35:1451–1468
Symington LS, Gautier J (2011) Double-strand break end resection and repair pathway choice. Annu Rev Genet 45:247–271
Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK (2015) GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33:187–197
Wang S, Zhang S, Wang W, Xiong X, Meng F, Cui X (2015) Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep 34:1473–1476
Wechter WP, Levi A, Harris KR, Davis AR, Fei Z, Katzir N, Giovannoni JJ, Salman-Minkov A, Hernandez A, Thimmapuram J, Tadmor Y, Portnoy V, Trebitsh T (2008) Gene expression in developing watermelon fruit. BMC Genom 9:275–288
Xie K, Zhang J, Yang Y (2014) Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol Plant 7:923–926
Xie K, Minkenberg B, Yang Y (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA 112:3570–3575
Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14:327
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2:1565–1572
Yu TA, Chiang CH, Wu HW, Li CM, Yang CF, Chen JH, Chen YW, Yeh SD (2011) Generation of transgenic watermelon resistant to Zucchini yellow mosaic virus and Papaya ringspot virus type W. Plant Cell Rep 30:359–371
Zhang B, Yang X, Yang C, Li M, Guo Y (2016) Exploiting the CRISPR/Cas9 system for targeted genome mutagenesis in Petunia. Sci Rep 6:20315
Zhou H, Liu B, Weeks DP, Spalding MH, Yang B (2014) Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res 42:10903–10914
Acknowledgements
We thank Dr. Shuhua Yang for technical help. We thank Dr. Qijun Chen for providing the CRIPSR/Cas9 genome targeting vector. This work was supported by grants from National Natural Science Foundation of China (31361140355, 31401893, 31272184, 31471785 and 31301738), Beijing Ministry of Science and Technology of China (2014BAD01B09), Beijing Municipal Science and Technology Commission of China (Z161100004916081, 6141001, 2014000021223TD03), Beijing Scholar Program (BSP026), Ministry of Agriculture of China (CARS-26) and BAAFS Innovation Team (JNKYT301601).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by N. Stewart.
S. Tian and L. Jiang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tian, S., Jiang, L., Gao, Q. et al. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep 36, 399–406 (2017). https://doi.org/10.1007/s00299-016-2089-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-016-2089-5