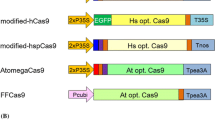
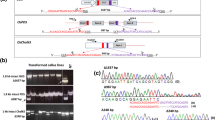
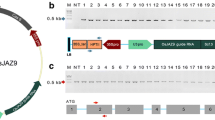
Abstract
Key message
Frequency of CRISPR/Cas9-mediated targeted mutagenesis varies depending on Cas9 expression level and culture period of rice callus.
Abstract
Recent reports have demonstrated that the CRISPR/Cas9 system can function as a sequence-specific nuclease in various plant species. Induction of mutation in proliferating tissue during embryogenesis or in germline cells is a practical means of generating heritable mutations. In the case of plant species in which cultured cells are used for transformation, non-chimeric plants can be obtained when regeneration occurs from mutated cells. Since plantlets are regenerated from both mutated and non-mutated cells in a random manner, any increment in the proportion of mutated cells in Cas9- and guide RNA (gRNA)-expressing cells will help increase the number of plants containing heritable mutations. In this study, we examined factors affecting mutation frequency in rice calli. Following sequential transformation of rice calli with Cas9- and gRNA- expression constructs, the mutation frequency in independent Cas9 transgenic lines was analyzed. A positive correlation between Cas9 expression level and mutation frequency was found. This positive relationship was observed regardless of whether the transgene or an endogenous gene was used as the target for CRISPR/Cas9-mediated mutagenesis. Furthermore, we found that extending the culture period increased the proportion of mutated cells as well as the variety of mutations obtained. Because mutated and non-mutated cells might proliferate equally, these results suggest that a prolonged tissue culture period increases the chance of inducing de novo mutations in non-mutated cells. This fundamental knowledge will help improve systems for obtaining non-chimeric regenerated plants in many plant species.




Similar content being viewed by others
Abbreviations
- CAPS:
-
Cleaved amplified polymorphic sequences
- Cas9:
-
CRISPR-associated endonuclease 9
- CDKB2:
-
Cyclin-Dependent Kinase B2
- CIM:
-
Callus induction medium
- CRISPR:
-
Clustered regularly interspaced short palindromic repeat
- gRNA:
-
Guide RNA
- HMF:
-
Highest mutation frequency
- NLS:
-
Nuclear localization signal
- nt:
-
Nucleotide
- PAM:
-
Protospacer adjacent motif
- RT-PCR:
-
Reverse transcriptase-polymerase chain reaction
- RC:
-
Ratio of calli with mutations
- YSA:
-
Young seeding albino
References
Ansai S, Kinoshita M (2014) Targeted mutagenesis using CRISPR/Cas system in medaka. Biol Open 3:362–371
Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V (2013) Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Method 9:39
Cho SW, Kim S, Kim JM, Kim JS (2013) Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31:230–232
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Endo M, Mikami M, Toki S (2015) Multigene Knockout Utilizing off-Target Mutations of the CRISPR/Cas9 System in Rice. Plant Cell Physiol 56:41–47
Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79:348–359
Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu JK (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23:1229–1232
Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L, Zeng L, Liu X, Zhu JK (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Nat Acad Sci U S A 111:4632–4637
Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32:279–284
Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31:227–229
Hyun Y, Kim J, Cho SW, Choi Y, Kim JS, Coupland G (2015) Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta 241:271–284
Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA (2013a) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239
Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013b) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41:e188
Jiang W, Yang B, Weeks DP (2014) Efficicent CRISPR/Cas9-mediated Gene Editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS One 9:e99225
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J (2013) RNA-programmed genome editing in human cells. Elife 2:e00471
Kuroda M, Kimizu M, Mikami C (2010) A simple set of plasmids for the production of transgenic plants. Biosci Biotechnol Biochem 74:2348–2351
Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31:688–691
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826
Mao Y, Zhang H, Xu N, Zhang B, Gao F, Zhu JK (2013) Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant 6:2008–2011
Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu LJ (2013) Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res 23:1233–1236
Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31:691–693
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, Gao C (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31:686–688
Sugano S, Shirakawa M, Takagi J, Matsuda Y, Shimada T, Hara-Nishimura I, Kohchi T (2014) CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol 55:475–481
Toki S (1997) Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol Biol Rep 15:16–21
Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47:969–976
Wang MB, Helliwell CA, Wu LM, Waterhouse PM, Peacock WJ, Dennis ES (2008) Hairpin RNAs derived from RNA polymerase II and polymerase III promoter-directed transgenes are processed differently in plants. RNA 14:903–913
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153:910–918
Xie K, Yang Y (2013) RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant 6:1975–1983
Xu R, Li H, Qin R, Wang L, Li L, Wei P, Yang J (2014) Gene targeting the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice 7:5
Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, Zhu JK (2014) The CRISPR/Cas9 system produced specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12:797–807
Zhou H, Liu B, Weeks DP, Spalding MH, Yang B (2014) Large chromosomal deletions heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res 42:10903–10914
Acknowledgments
We thank K. Amagai, R. Aoto, A. Nagashii and F. Suzuki for general experimental technical support. This research was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation PGE1001) and the NIAS Strategic Research Fund. This work was also supported by the Council for Science, Technology and Innovation (CSTI), Cross-ministerial Strategic Innovation Promotion Program (SIP), “Technologies for creating next-generation agriculture, forestry and fisheries” (funding agency: Bio-oriented Technology Research Advancement Institution, NARO).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by K. Toriyama.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mikami, M., Toki, S. & Endo, M. Parameters affecting frequency of CRISPR/Cas9 mediated targeted mutagenesis in rice. Plant Cell Rep 34, 1807–1815 (2015). https://doi.org/10.1007/s00299-015-1826-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-015-1826-5