Abstract
Key message
A putative RNA-binding protein with a single RNA Recognition Motif (At3G63450) is involved in anthocyanin biosynthesis via its ability to modulate the transcript level of a major positive regulator PAP1 in Arabidopsis.
Abstract
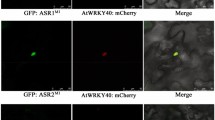
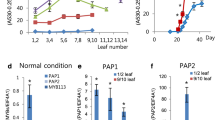
The R2R3 MYB-activator production of anthocyanin pigment 1 (PAP1)/MYB75 plays a major role in anthocyanin biosynthesis in Arabidopsis in combination with one of three bHLH activators including transparent test 8 (TT8), enhancer of glabra3 (EGL3), glabra3 (GL3), and the WD-repeat transcription factor transparent testa 1 (TTG1), forming ternary MYB-basic HLH-WD40 complexes. Transcriptional activation of PAP1 expression is largely triggered by changes in light color and intensity, temperature fluctuations, nutrient status, and sugar and hormone treatments. However, the immediate upstream and downstream regulatory factors for PAP1 transcription are largely unknown. In the present study, using a T-DNA insertional mutagenesis approach, we transformed pap1-Dominant (pap1D) plants to modulate the levels of endogenous PAP1 transcripts. We employed Restriction Site Extension (RSE)-PCR analysis of 247 homogenous T3 genetic mutant lines exhibiting variations in anthocyanin accumulation compared to pap1D and identified 92 lines with T-DNA integrated in either intra- or inter-genic locations. This analysis revealed 80 novel candidate proteins, including a putative RNA-binding protein with a single RNA Recognition Motif (At3G63450), which may directly or indirectly regulate PAP1 expression at the transcriptional level.




Similar content being viewed by others
References
Allan AC, Hellens RP, Laing WA (2008) MYB transcription factors that colour our fruit. Trends Plant Sci 13:99–102
Borevitz JO, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12:2383–2394
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Dare A, Schaffer RJ, Lin-Wang K, Allan C, Hellens RP (2008) Identification of a cis-regulatory element by transient analysis of coordinately regulated genes. Plant Methods 4:17
Das PK, Shin DH, Choi SB, Yoo SD, Choi G, Park Y-I (2012a) Cytokinins enhance sugar-induced anthocyanin biosynthesis in Arabidopsis. Mol Cells 34:93–101
Das PK, Shin DH, Choi SB, Park Y-I (2012b) Sugar-hormone cross-talk in anthocyanin biosynthesis. Mol Cells 34:501–507
Daxinger L, Hunter B, Sheikh M, Jauvion V, Gasciolli V, Vaucheret H, Matzke M, Furner I (2008) Unexpected silencing effects from T-DNA tags in Arabidopsis. Trends Plant Sci 13:4–6
Devaiah BN, Nagarajan VK, Raghothama KG (2007) Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol 145:147–159
Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55:940–953
Forsbach A, Schubert D, Lechtenberg B, Gils M, Schmidt R (2003) A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. Plant Mol Biol 52:161–176
Frishman D, Mokrejs M, Kosykh D, Kastenmüller G, Kolesov G, Zubrzycki I, Gruber C, Geier B, Kaps A, Albermann K, Volz A, Wagner C, Fellenberg M, Heumann K, Mewes HW (2003) The PEDANT genome database. Nucleic Acids Res 31:207–211
Gao Y, Zhao Y (2013) Epigenetic suppression of T-DNA insertion mutants in Arabidopsis. Mol Plant 6:539–549
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53:814–827
Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23:1512–1522
Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D (2005) Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17:92–102
Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK, Ulm R (2010) Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci USA 16:20132–20137
Hodgkin J (2005) Genetic suppression. WormBook 27:1–13
Jeong SW, Das PK, Jeoung SC, Song JY, Lee HK, Kim YK, Kim WJ, Park YI, Yoo SD, Choi SB, Choi G, Park Y-I (2010) Ethylene suppression of sugar-induced anthocyanin pigmentation in Arabidopsis. Plant Physiol 154:1514–1531
Ji J, Braam J (2010) Restriction site extension PCR: a novel method for high throughput characterization of target DNA fragments and genome walking. PLoS One 5:e10577
Kang H, Park SJ, Kwak KJ (2013) Plant RNA chaperones in stress response. Trends Plant Sci 18:100–106
Kliebenstein DJ, Lim JE, Landry LG, Last RL (2002) Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation. Plant Physiol 130:234–243
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10:236–242
Koh EJ, Kwon YR, Kim KI, Hong SW, Lee H (2009) Altered ARA2 (RABA1a) expression in Arabidopsis reveals the involvement of a Rab/YPT family member in auxin-mediated responses. Plant Mol Biol 70:113–122
Koops P, Pelser S, Ignatz M, Klose C, Marrocco-Selden K, Kretsch T (2011) EDL3 is an F-box protein involved in the regulation of abscisic acid signaling in Arabidopsis thaliana. J Exp Bot 62:5547–5560
Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11:2283–2290
Liu Z, Shi MZ, Xie DY (2014) Regulation of anthocyanin biosynthesis in Arabidopsis thaliana red pap1-D cells metabolically programmed by auxins. Planta 239:765–781
Lorković ZJ (2009) Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci 14:229–236
Matsui K, Umemura Y, Ohme-Takagi M (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55:954–967
Matzke MA, Priming M, Trnovsky J, Matzke AJM (1989) Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J 8:643–649
Mayerhofer R, Koncz-Kalman Z, Nawrath C, Bakkeren G, Crameri A, Angelis K, Redei GP, Schell J, Hohn B, Koncz C (1991) T-DNA integration: a mode of illegitimate recombination in plants. EMBO J 10:697–704
Ni DA, Sozzani R, Blanchet S, Domenichini S, Reuzeau C, Cella R, Bergounioux C, Raynaud C (2009) The Arabidopsis MCM2 gene is essential to embryo development and its over-expression alters root meristem function. New Phytol 184:311–322
Petroni K, Tonelli C (2011) Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci 181:219–229
Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A (1998) The 1.7 Å crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature 5:97–101
Revenkova E, Masson J, Koncz C, Afsar K, Jakovleva L, Paszkowski J (1999) Involvement of Arabidopsis thaliana ribosomal protein S27 in mRNA degradation triggered by genotoxic stress. EMBO J 18:490–499
Rosado A, Schapire AL, Bressan RA, Harfouche AL, Hasegawa PM, Valpuesta V, Botella MA (2006) The Arabidopsis tetratricopeptide repeat-containing protein TTL1 is required for osmotic stress responses and abscisic acid sensitivity. Plant Physiol 142:1113–1126
Rowan DD, Cao M, Lin-Wang K, Cooney JM, Jensen DJ, Austin PT, Hunt MB, Norling CR, Hellens P, Schaffer RJ, Allan AC (2009) Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol 182:102–115
Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR (2009) Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21:3567–3584
Shin DH, Choi MG, Kim K, Bang G, Cho M, Choi SB, Choi G, Park Y-I (2013a) HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett 587:1543–1547
Shin DH, Choi MG, Lee HK, Cho M, Choi SB, Choi G, Park YI (2013b) Calcium dependent sucrose uptake links sugar signaling to anthocyanin biosynthesis in Arabidopsis. Biochem Biophys Res Commun 430:634–639
Stevens R, Mariconti L, Rossignol P, Perennes C, Cella R, Bergounioux C (2002) Two E2F sites in the Arabidopsis MCM3 promoter have different roles in cell cycle activation and meristematic expression. J Biol Chem 277:32978–32984
Tanaka Y, Ohmiya A (2008) Seeing is believing: engineering anthocyanin and carotenoid biosynthetic pathways. Curr Opin Biotechnol 19:190–197
Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139:1840–1852
Wang X, Chen Y, Zou J, Wu W (2007) Involvement of a cytoplasmic glyceraldehyde-3-phosphate dehydrogenase GapC-2 in low-phosphate-induced anthocyanin accumulation in Arabidopsis. Chin Sci Bull 52:1764–1770
Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5:218–223
Yoshida H, Nagata M, Saito K, Wang KL, Ecker JR (2005) Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol 5:14
Acknowledgments
This work was supported by Grant PJ8205 from the Next-Generation BioGreen 21 Program, Rural Development Administration, and Grant 2011-0031343 from the Advanced Biomass Research and Development Center, Republic of Korea.
Conflict of interest
The authors have no competing financial interests to declare.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. S. Shin.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shin, D.H., Cho, M., Choi, M.G. et al. Identification of genes that may regulate the expression of the transcription factor production of anthocyanin pigment 1 (PAP1)/MYB75 involved in Arabidopsis anthocyanin biosynthesis. Plant Cell Rep 34, 805–815 (2015). https://doi.org/10.1007/s00299-015-1743-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-015-1743-7